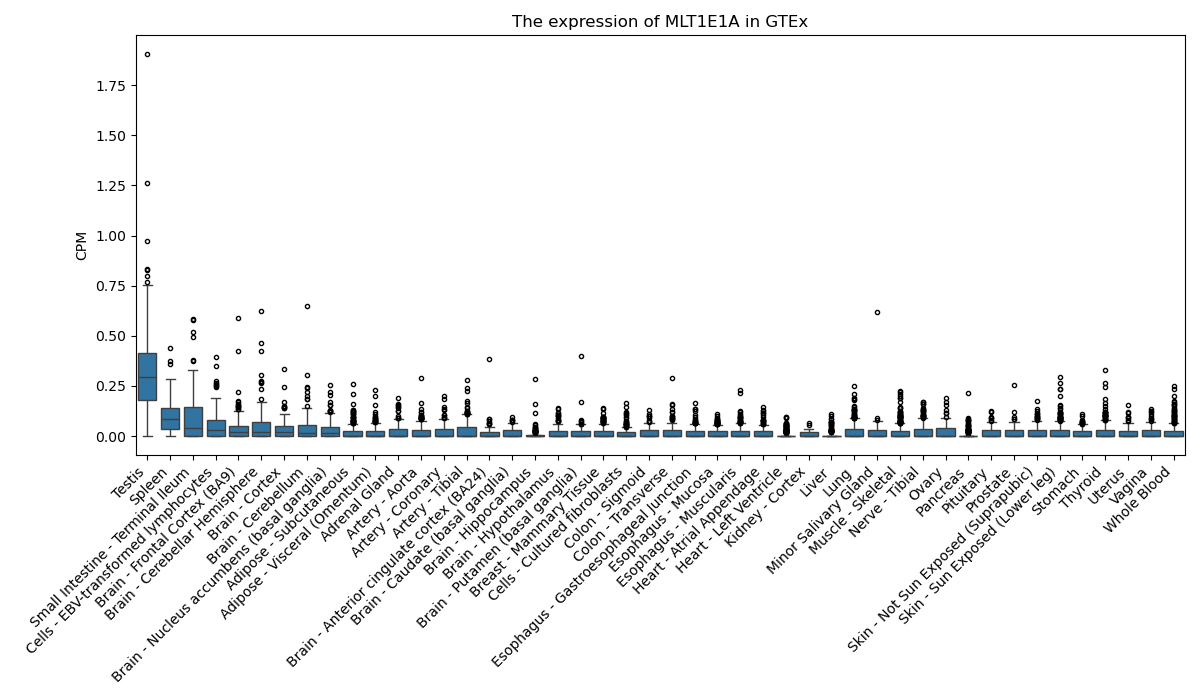
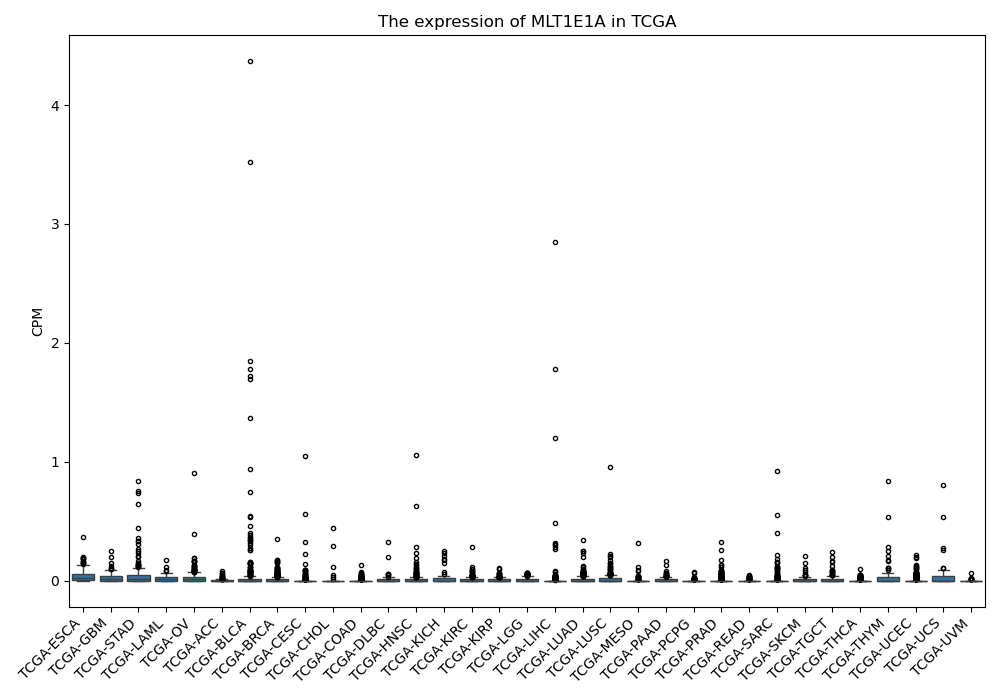
MLT1E1A
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0001128 |
|---|---|
| TE superfamily | MaLR |
| TE class | LTR |
| Species | Eutheria |
| Length | 680 |
| Kimura value | 23.96 |
| Tau index | 0.9809 |
| Description | MLT1E1A Long Terminal Repeat for ERVL-MaLR retrotransposon |
| Comment | Note that a full-length ERV-MaLR (Mammalian apparent LTR retrotransposon) element is derived from an ERV (Endogenous Retrovirus), but only contains the Gag gene (that is, no Pol or Env). |
| Sequence |
TGTGGTAGGCAGAATTCTAAGATGGCCCCCAAGATTCCCGCCCCCTGGTGTACACGCCCTGTATAATCCCCTCCCCTTGAGTGTGGGCGGGACCTGTGAATATGATGGGATATCACTCCCGTGATTAGGTTACATTATATGGCAAAGGTGAAGGGATTTTGCAGATGTAATTAAGGTCCCTAATCAGTTGACTTTGAGTTAATCAAAAGGGAGATTATCCTGGGTGGGCCTGACCTAATCAGGTGAGCCCTTAAAAGAGGCATGGGCCCTCCAGAGAGAAGAACAGAGAGATTCTCCTGCTGGCCTTGAAGAAGCAAGCTGCCATGTTGTGAGAGGGCCTATGGAGAGGGCCACGTGGCAAGGANCTGNGGGCGGCCTCTAGGAGCTGAGAGCGGCCCCCGGCCGACAGCCAGCAAGAAAACGGGGACCTCAGTCCTACAGCCGCAAGGAANTGAATTCTGCCAACAACCTGAATGAGCTTGGAAGNGGACCCTAAGCCTCAGATGAGAACGCAGCCCTAGCCGACACCTTGATTNCAGCCTTGTGAGACCCTGAGCAGAGGACCCAGCTAAGCCGTGCCCGGACTCCTGACCCACAGAAACTGTGAGATAATAAATGTGTGTTGTTTTAAGCCGCTAAGTTTGTGGTAATTTGTTACGCAGCAATAGAAAACTAATACA
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| MLT1E1A | BZIP68 | 351 | 360 | - | 15.97 | TGCCACGTGG |
| MLT1E1A | WRKY29 | 187 | 195 | - | 15.92 | AAAGTCAAC |
| MLT1E1A | SP4 | 68 | 76 | - | 15.85 | GGGGAGGGG |
| MLT1E1A | ZNF667 | 244 | 254 | - | 15.80 | TTAAGGGCTCA |
| MLT1E1A | CTCFL | 40 | 47 | - | 15.72 | CAGGGGGC |
| MLT1E1A | WRKY45 | 187 | 196 | - | 15.71 | CAAAGTCAAC |
| MLT1E1A | SP2 | 36 | 44 | - | 15.71 | GGGGCGGGA |
| MLT1E1A | ci | 220 | 230 | - | 15.66 | GGCCCACCCAG |
| MLT1E1A | btd | 37 | 45 | - | 15.62 | GGGGGCGGG |
| MLT1E1A | WRKY14 | 186 | 195 | - | 15.61 | AAAGTCAACT |
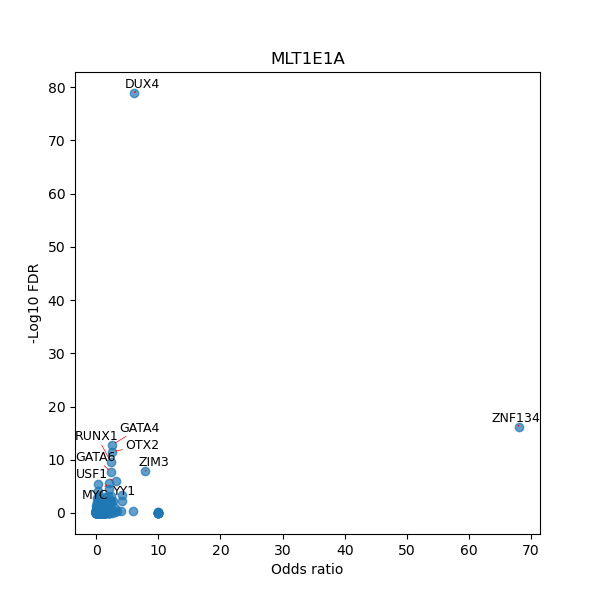
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.