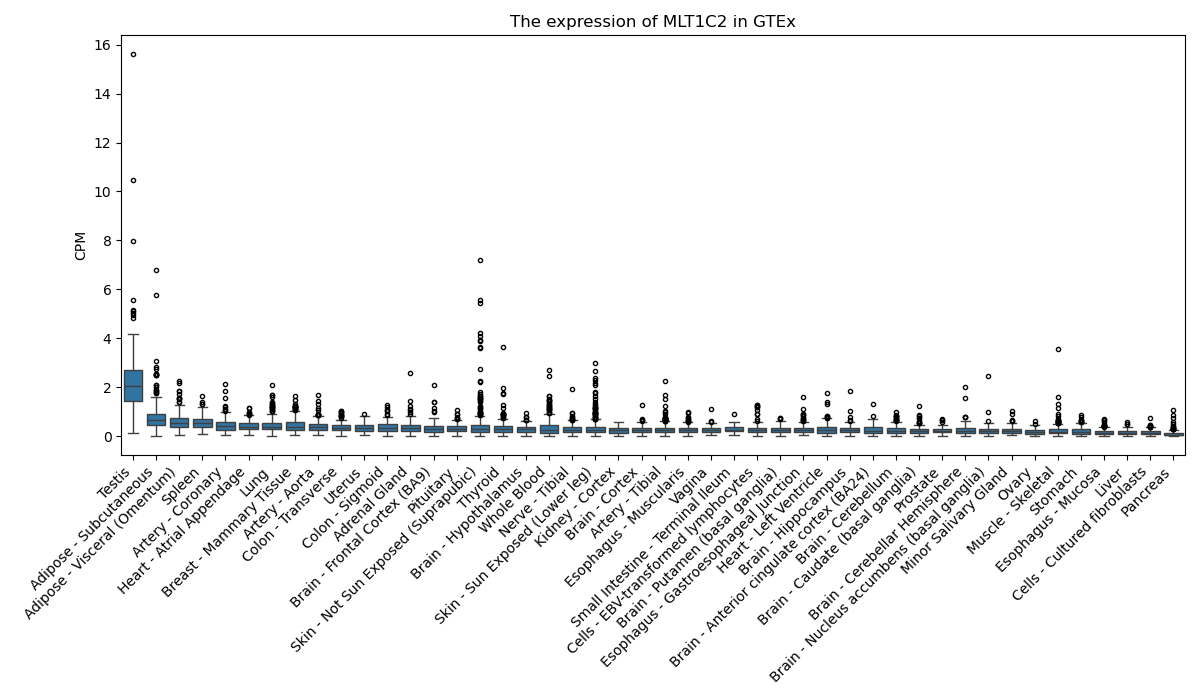
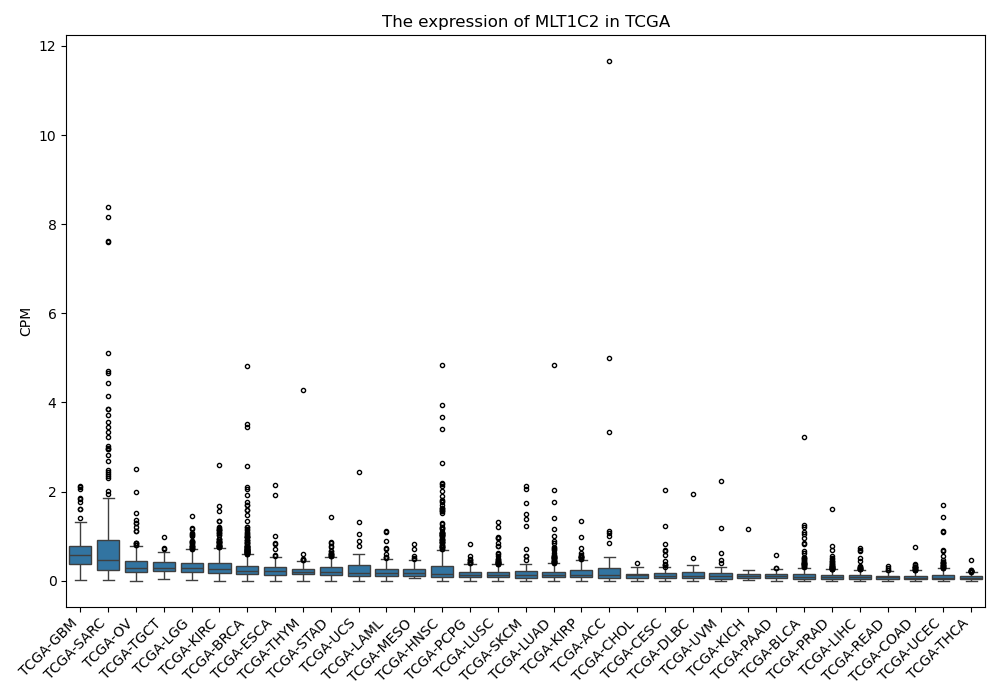
MLT1C2
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0001221 |
|---|---|
| TE superfamily | MaLR |
| TE class | LTR |
| Species | Eutheria |
| Length | 461 |
| Kimura value | 21.32 |
| Tau index | 0.8666 |
| Description | Mammalian long terminal repeat (MLT1C subfamily) |
| Comment | Specific to boreoeutherians. |
| Sequence |
TGTTGTGGGTTGAATTGTGTCCCCCAAAAAGATATGTTCAAGTCCTAACCCCCGGNACCTGTGAATGTGACCTTATTTGGAAATAGGGTCTTTGCAGATGTAATCAAGTTAAGATGAGGTCATACTGGATTAGGGTGGGCCCTAATCCAATGACTGGTGTCCTTATAAGAAGAGGGAAATNTGGACACAGAGACACACAGGGAGAAGGCCATGTGAAGACGGAGGCAGAGATTGGAGTGATGCGGCTACAAGCCAAGGAACGCCAAGGATTGCCGGCAACCACCAGAAGCTAGGAAGAGGCAAGGAAGGATTCTCCCCTAGAGCCTTCAGAGGGAGCACGGCCCTGCCGACACCTTGATTTCGGACTTCTGGCCTCCAGAACTGTGAGAGAATAAATTTCTGTTGTTTTAAGCCACCCAGTTTGTGGTANTTTGTTACGGCAGCCCTAGGAAACTAATACA
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| MLT1C2 | Gfi1B | 225 | 234 | - | 14.07 | CAATCTCTGC |
| MLT1C2 | TCP19 | 135 | 142 | - | 13.97 | GGGCCCAC |
| MLT1C2 | TCP4 | 135 | 142 | - | 13.94 | GGGCCCAC |
| MLT1C2 | AGL6 | 65 | 83 | - | 13.94 | TTTCCAAATAAGGTCACAT |
| MLT1C2 | MCM1 | 71 | 82 | + | 13.84 | CCTTATTTGGAA |
| MLT1C2 | ZIPIC | 440 | 449 | + | 13.83 | GCAGCCCTAG |
| MLT1C2 | AGL13 | 66 | 83 | - | 13.83 | TTTCCAAATAAGGTCACA |
| MLT1C2 | nhr-70 | 243 | 251 | - | 13.81 | TTGTAGCCG |
| MLT1C2 | ERF6 | 271 | 277 | + | 13.72 | TGCCGGC |
| MLT1C2 | ERF6 | 272 | 278 | - | 13.72 | TGCCGGC |
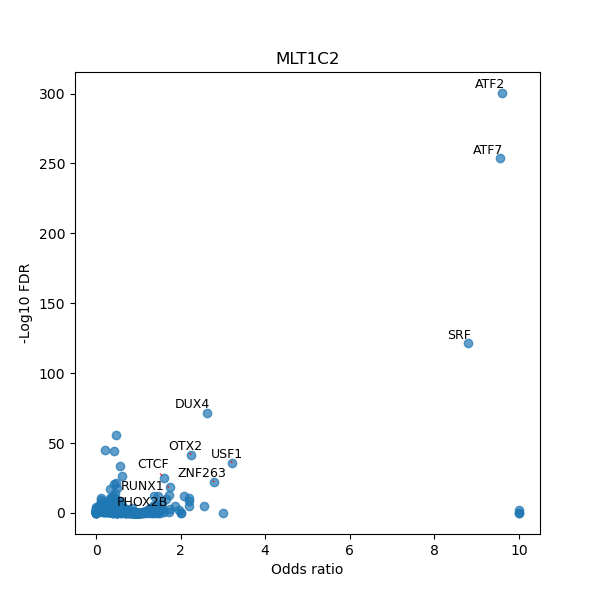
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.