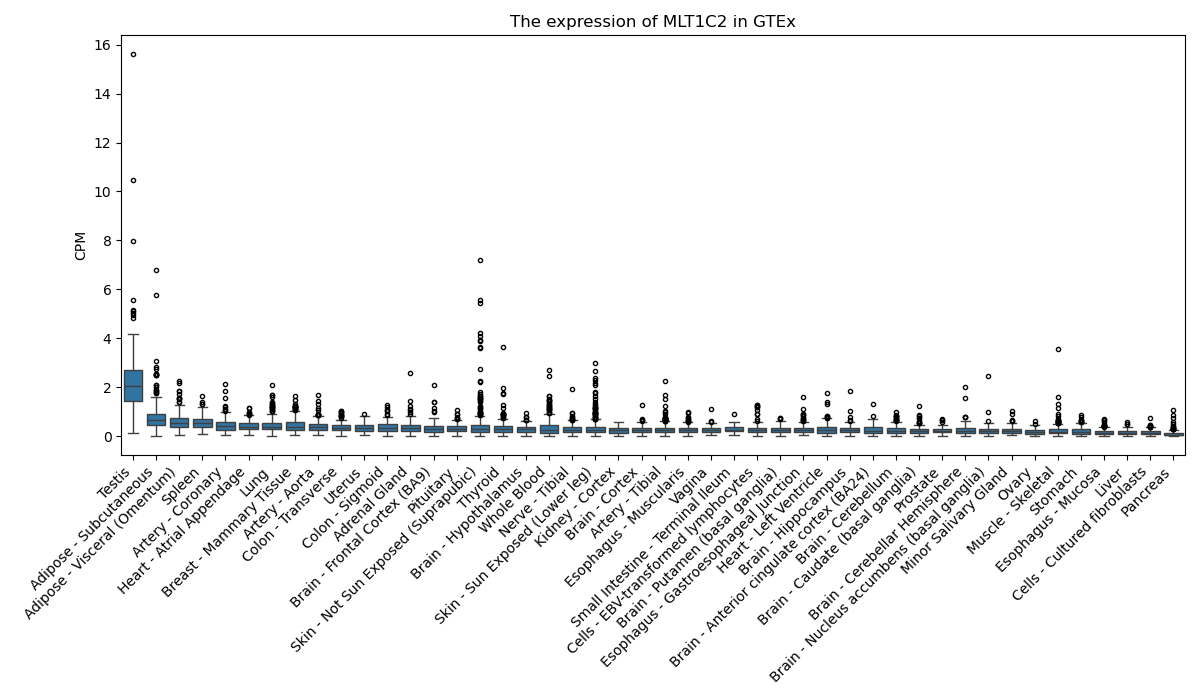
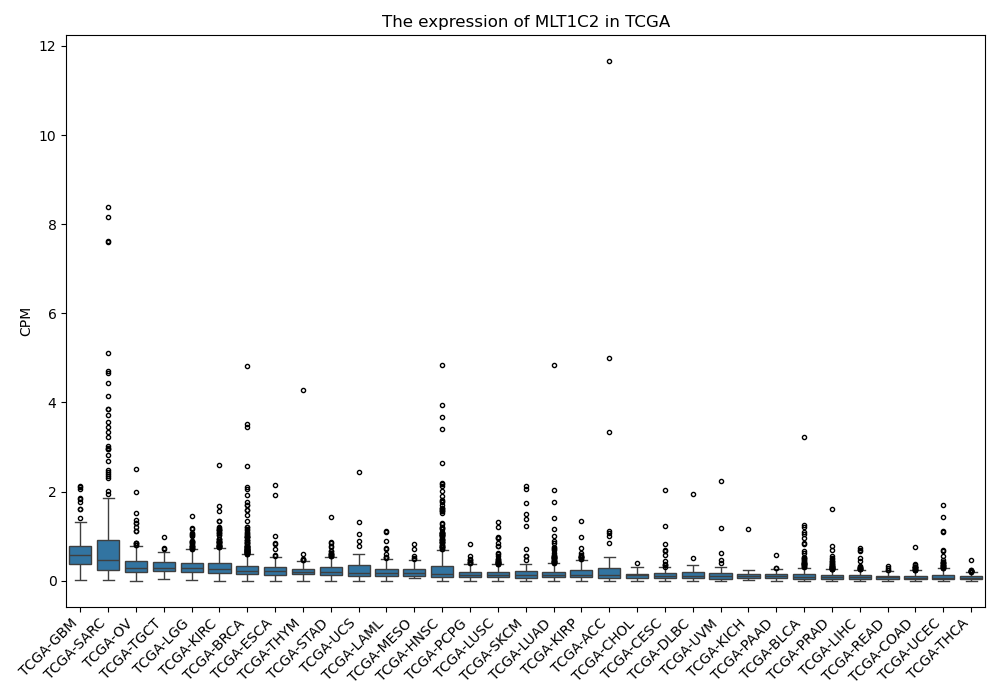
MLT1C2
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0001221 |
|---|---|
| TE superfamily | MaLR |
| TE class | LTR |
| Species | Eutheria |
| Length | 461 |
| Kimura value | 21.32 |
| Tau index | 0.8666 |
| Description | Mammalian long terminal repeat (MLT1C subfamily) |
| Comment | Specific to boreoeutherians. |
| Sequence |
TGTTGTGGGTTGAATTGTGTCCCCCAAAAAGATATGTTCAAGTCCTAACCCCCGGNACCTGTGAATGTGACCTTATTTGGAAATAGGGTCTTTGCAGATGTAATCAAGTTAAGATGAGGTCATACTGGATTAGGGTGGGCCCTAATCCAATGACTGGTGTCCTTATAAGAAGAGGGAAATNTGGACACAGAGACACACAGGGAGAAGGCCATGTGAAGACGGAGGCAGAGATTGGAGTGATGCGGCTACAAGCCAAGGAACGCCAAGGATTGCCGGCAACCACCAGAAGCTAGGAAGAGGCAAGGAAGGATTCTCCCCTAGAGCCTTCAGAGGGAGCACGGCCCTGCCGACACCTTGATTTCGGACTTCTGGCCTCCAGAACTGTGAGAGAATAAATTTCTGTTGTTTTAAGCCACCCAGTTTGTGGTANTTTGTTACGGCAGCCCTAGGAAACTAATACA
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| MLT1C2 | Rarg | 106 | 122 | + | 3.82 | AAGTTAAGATGAGGTCA |
| MLT1C2 | Atf-2 | 113 | 123 | + | 3.36 | GATGAGGTCAT |
| MLT1C2 | Atf-2 | 114 | 124 | - | 1.95 | TATGACCTCAT |
| MLT1C2 | TGIF1 | 151 | 162 | + | 1.59 | TGACTGGTGTCC |
| MLT1C2 | EWSR1-FLI1 | 292 | 309 | + | -0.28 | AGGAAGAGGCAAGGAAGG |
| MLT1C2 | Rarb | 106 | 122 | + | -2.23 | AAGTTAAGATGAGGTCA |
| MLT1C2 | Rarg | 362 | 376 | - | -2.91 | GAGGCCAGAAGTCCG |
| MLT1C2 | EWSR1-FLI1 | 296 | 313 | + | -11.95 | AGAGGCAAGGAAGGATTC |
| MLT1C2 | BPC5 | 183 | 212 | + | -44.50 | GGACACAGAGACACACAGGGAGAAGGCCAT |
| MLT1C2 | BPC5 | 201 | 230 | + | -48.96 | GGAGAAGGCCATGTGAAGACGGAGGCAGAG |
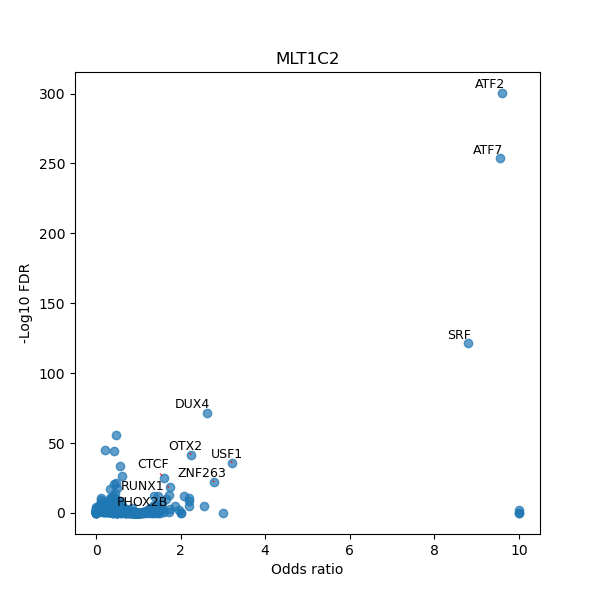
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.