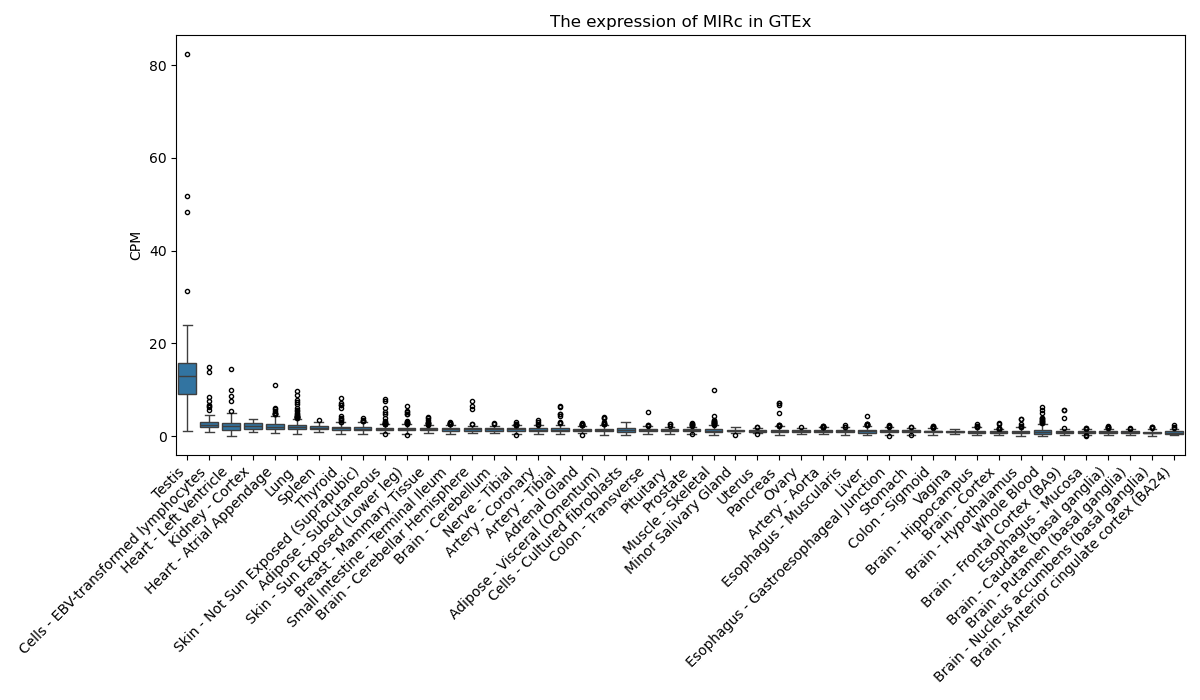
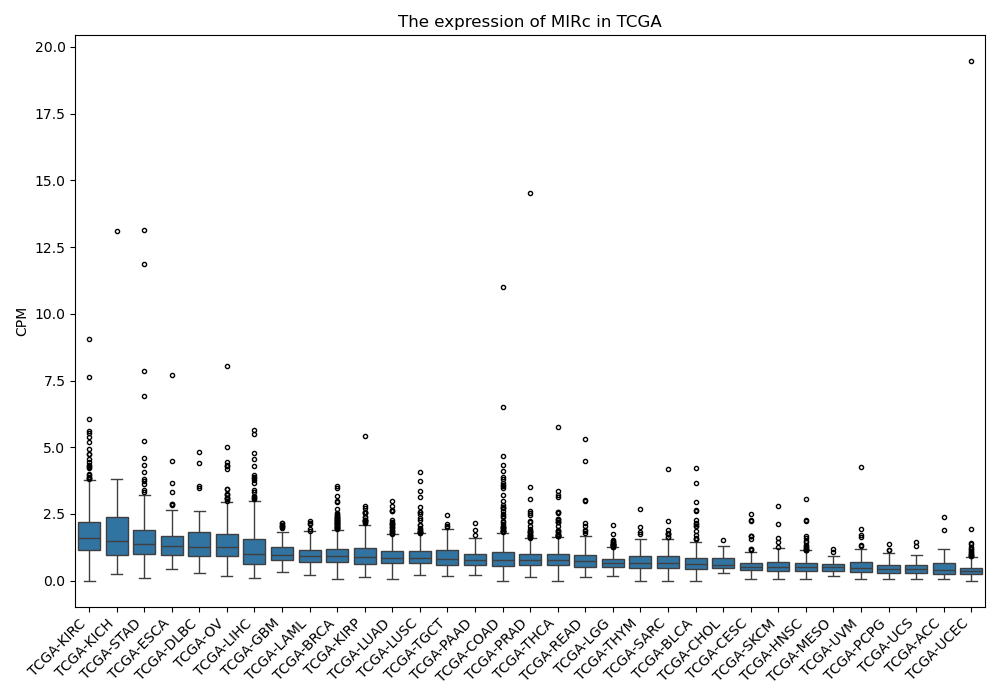
MIRc
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000976 |
|---|---|
| TE superfamily | L2-end |
| TE class | SINE |
| Species | Mammalia |
| Length | 268 |
| Kimura value | 35.35 |
| Tau index | 0.9012 |
| Description | MIRc (Mammalian-wide Interspersed Repeat - variant c) |
| Comment | MIRc is a pan-mammalian SINE with a 5' end derived from a tRNA, a central deeply-conserved CORE region (shared with other MIRs), and a 3' terminal ~55bp related to an L2 LINE. |
| Sequence |
CGAGGCAGTGTGGTGCAGTGGAAAGAGCACTGGACTTGGAGTCAGGAAGACCTGGGTTCGAGTCCCGGCTCTGCCACTTACTAGCTGTGTGACCTTGGGCAAGTCACTTAACCTCTCTGAGCCTCAGTTTCCTCATCTGTAAAATGGGGATAATAATACCTGCCCTGCCTACCTCACAGGGTTGTTGTGAGGATCAAATGAGATAATGTATGTGAAAGCGCTTTGTAAACTGTAAAGTGCTATACAAATGTAAGGNGTTATTATTATT
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| MIRc | PLAG1 | 112 | 125 | - | 4.97 | GAGGCTCAGAGAGG |
| MIRc | THRB | 118 | 135 | + | 4.86 | TGAGCCTCAGTTTCCTCA |
| MIRc | ZSCAN31 | 154 | 171 | + | 3.35 | TAATACCTGCCCTGCCTA |
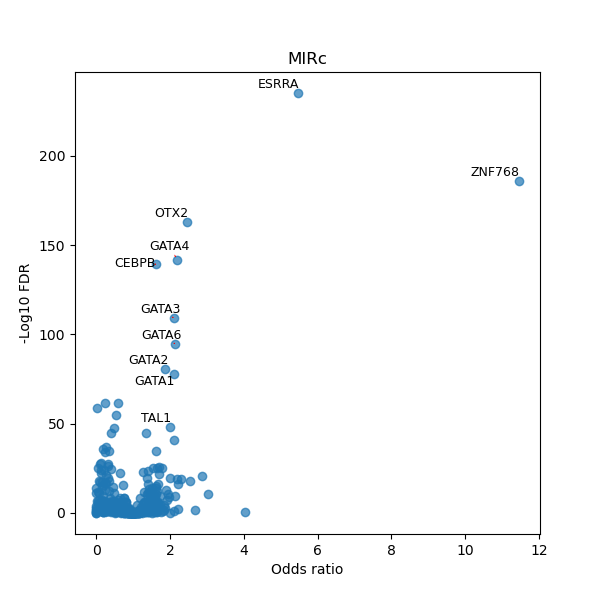
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.