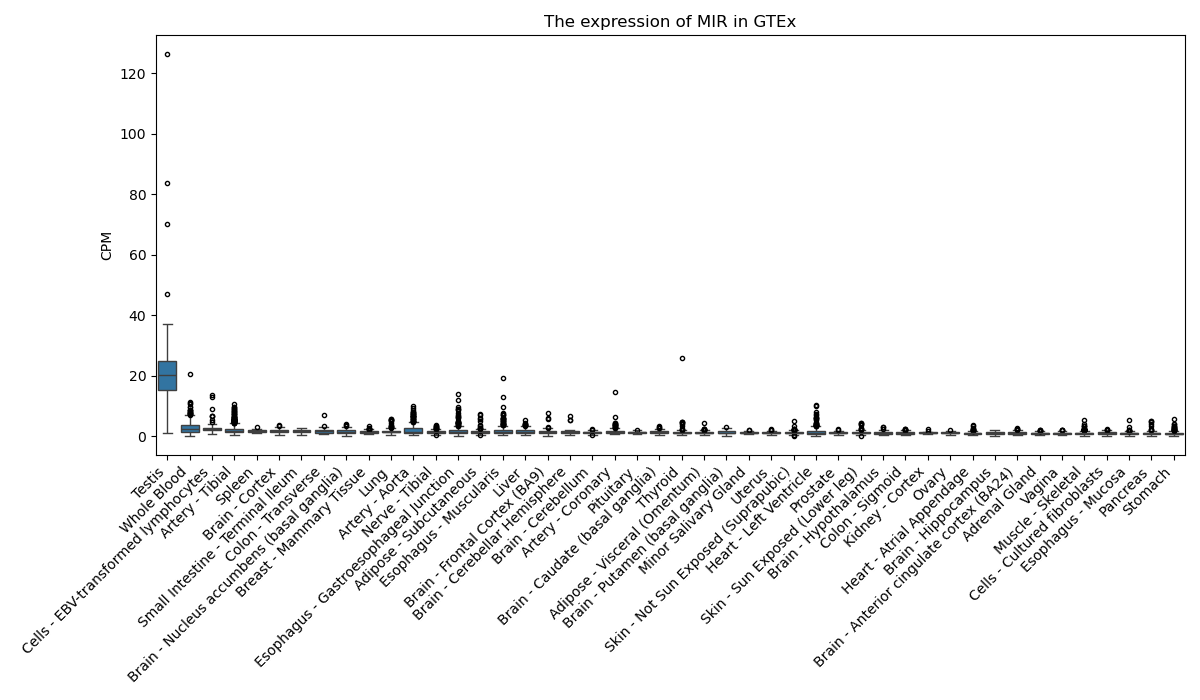
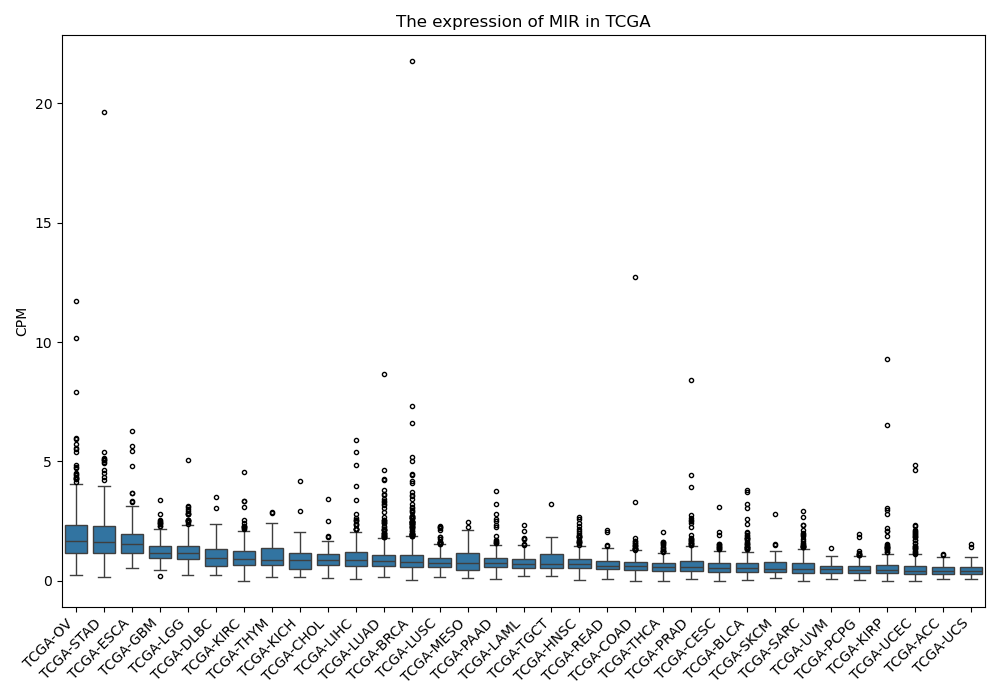
MIR
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000001 |
|---|---|
| TE superfamily | L2-end |
| TE class | SINE |
| Species | Mammalia |
| Length | 262 |
| Kimura value | 31.97 |
| Tau index | 0.9376 |
| Description | MIR (Mammalian-wide Interspersed Repeat) |
| Comment | MIR is a pan-mammalian SINE with a 5' end derived from a tRNA, a central deeply-conserved CORE region, and a 3' terminal ~55bp related to an L2 LINE. |
| Sequence |
ACAGTATAGCATAGTGGTTAAGAGCACGGGCTCTGGAGCCAGACTGCCTGGGTTCGAATCCCGGCTCTGCCACTTACTAGCTGTGTGACCTTGGGCAAGTTACTTAACCTCTCTGTGCCTCAGTTTCCTCATCTGTAAAATGGGGATAATAATAGTACCTACCTCATAGGGTTGTTGTGAGGATTAAATGAGTTAATACATGTAAAGCGCTTAGAACAGTGCCTGGCACATAGTAAGCGCTCAATAAATGTTAGCTATTATT
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| MIR | ESRRA | 86 | 94 | - | 15.37 | CCAAGGTCA |
| MIR | PK06182.1 | 156 | 163 | + | 15.27 | TACCTACC |
| MIR | Esrrg | 86 | 94 | - | 14.81 | CCAAGGTCA |
| MIR | ERR | 86 | 93 | - | 14.77 | CAAGGTCA |
| MIR | MYB3 | 156 | 163 | - | 14.67 | GGTAGGTA |
| MIR | ZNF768 | 107 | 115 | + | 14.62 | ACCTCTCTG |
| MIR | ESRRB | 85 | 94 | - | 14.41 | CCAAGGTCAC |
| MIR | pita | 51 | 68 | - | 14.17 | AGAGCCGGGATTCGAACC |
| MIR | Hr39 | 86 | 93 | - | 14.02 | CAAGGTCA |
| MIR | hr3 | 83 | 90 | - | 13.92 | GGTCACAC |
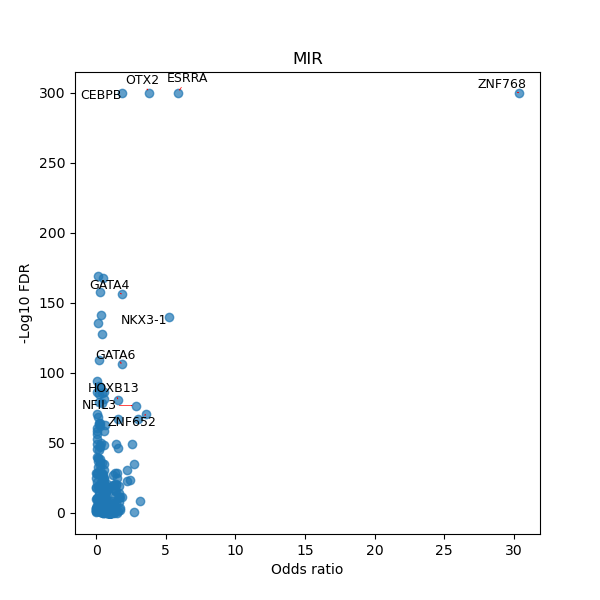
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.