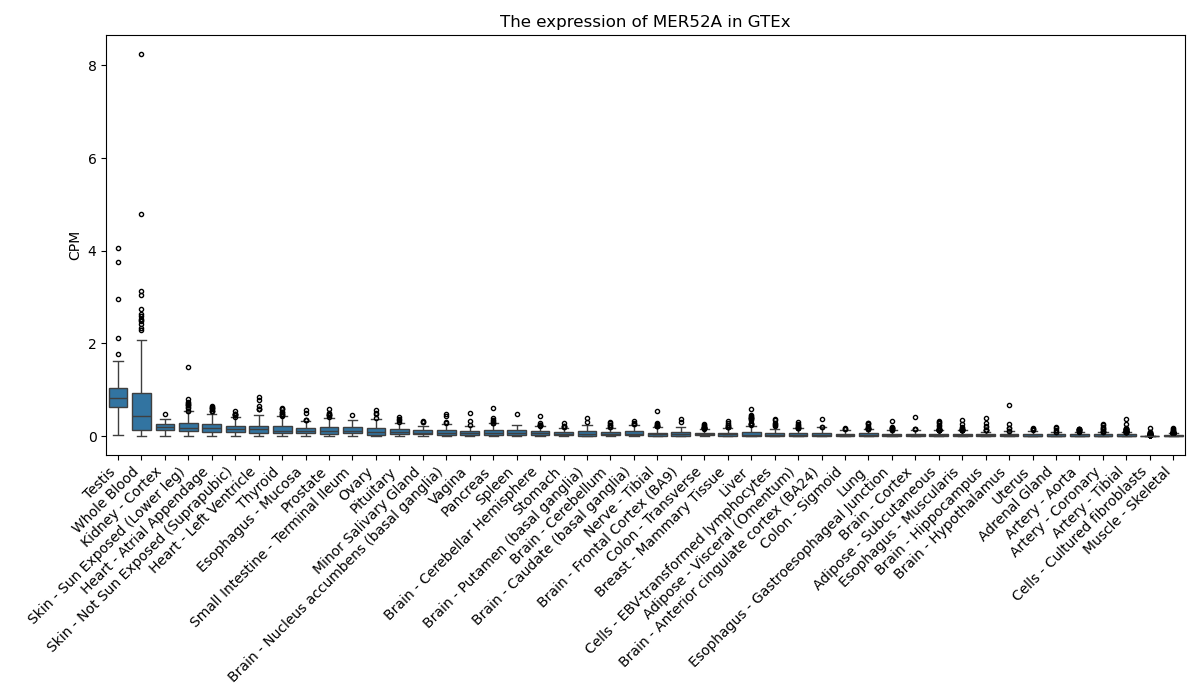
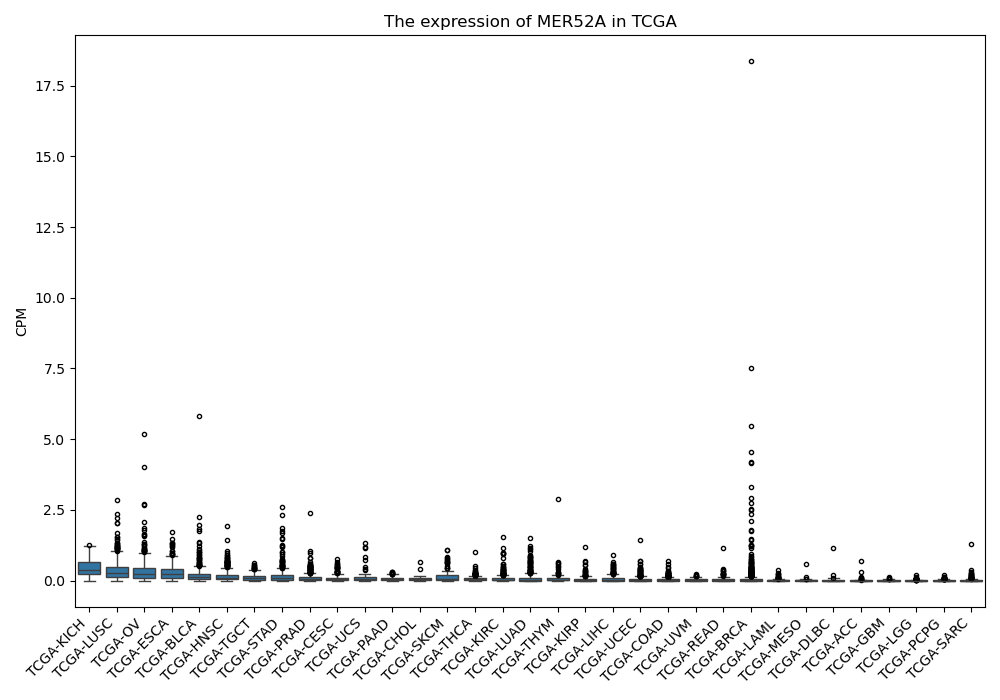
MER52A
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000856 |
|---|---|
| TE superfamily | ERV1 |
| TE class | LTR |
| Species | Haplorrhini |
| Length | 1755 |
| Kimura value | 12.01 |
| Tau index | 0.9189 |
| Description | MER52A Long Terminal Repeat for ERV1 endogenous retrovirus |
| Comment | MER52A is associated with MER52-int, and has 4 bp TSDs. The originally reported orientation has been reversed. Note that MER stands for MEdium Reiteration frequency interspersed repeat. As such, MERs as a group are a grab bag of DNA transposons and retrotransposons. |
| Sequence |
TGATGGCAGCGGCGGCCCGTCTGGAGCGGCCGCTGCNANGACGCCGGCTGCAGCGGGGGAGGCGCGGCCGGGGCTGCGCGCTCCACGGAGCCGGCGGGAGCCGGGAACAGGCGGGAGCCCCGCCCCCTTCCGAGTTGGGGCGGGAGCNNCCCGGGTGCGGCTGCAGCCGCCCANNCGCGGACCCGGGCNTCCCCGTGCTCNTGGGGNCCGGGAGCAGGCAGGAGCCCCGCCCTCCCGGGCGCAGCTGCAGCCGCCCAGCCGCGGCTGCGGACCCGGGCATCCCTGCGCTCTCGGGGGCCCGGGAAGNCCCCCCTGCCCCCGCAGGCTTGGAAGTGCCTGCTCCCGCCGCGNNCGNCCCCGCCCCGNNNNCGNCGNNCGNNCCGCAGGCTCGGAAGTGCCTGCTCCCGCTGCCTGGCCTCTCCCCGCTCCCGGCGCCCGCTCCGATTTCGGAGCAAAGTTGNGGCCGAGCCCGGGCGCTGTCGCGACCCGGCCGGGTGTGCGCGCGCTCGGGGCGGCGCTGACACGCCAGCCCCCTGCCGCCTCGGCCCCCTCCGGACTTTGGGCGCCGACGAGCACGGGAGGGAGGCCGAGGGGGGGCTGAGGGCGGCTCGGCGCGGGCCTGCAGGCGCCCCTCGGCACGAACAGCCTGGGCGCCATGGACGGCAGCAGGAGGCAGACAGGCTCCTGGGCGGAAAGGGGCGGGTCCCCGGTGAAGCCCCACCTTCAAGCCAGGGACGGCCTGAAGCCTGGGGGCCGGGCTGCCAGTTCCGGGTGGAGTCCGCGGACCGGAGTGAGAACTTATGGTGCTTTTTCCGGGCCCGCCCATGGCCGCCCATGGACCAATCAGCACGCACTTCCTCCCCTCTGAAGCCCATAAAAACCCCGGACTCAGCCAGACTCGGGCAGACGTCGGGACGACCAGCTGCGGAGAGGAGCTACCCACTCCGGGNTCTCCTCTCTGCTGAGAGCTGNANAGNCGTCGGGACGACCTGCCTGCGGAGAGGAGCTACCCACTCCAGGGTCTCCTCTCTGCTGAGAGCTGAACACTCGTCGGGACGACCTGCCTGCGGAAAGGAGCTACCCACTNCGGGTCTCCTGNGAGCTGTTCTGTCGCTCAATAAAGCTCCTCTTCGCCTTGCTCACCCTCCANTTGTCCGCGTACCTCATTCTTCCTGGACGCGGGACAAGAACTCGGGACCCGCCGAATGGCGGGGCTGAAAGAGCTGTAACACAAACAGGGCTGAAACACGCCCCCTGCTCGCCACGTTGCGGGCGACGAGAAGGAGAGAAGAGCTGCGGCCCTTCGGGGAGCCCAGACCTAGGAGCTCCCCGAGCCAGGGCTGTGACACCCTCTTTGGGGCTCTGCGGTTCCTGGCGTCTCCAAGCTTCCGGGCGCCACCGCGTTCCCCGGTGCCCGCNGTGGAAGCCGCTTGCGGTACGCCTGGTCCAGCCGCAGCCTCGCAGGGAGCCGGCGCCCGTGCCGGCGCCTGGAGCTGCCCGCCCCGCCGCAGCCGGCGTGCCTGGCTGTGCGCAGTGGCCGGACCCCGCGCTCGCTCGCTCACACACCCCTCGCCGCTCCGCGCCTGGCTCGCCCTTGGCAGGCGTGGGATCCGGGCCGGTAGCGCGAGCCGAGCGCAGCCTGCCAGGCCGAGTGGGCGGAACGAGCCCAGCGGGCCCGAGCAAAACTCGGGCAAAGGCGCCACCGGCCACAGAGGTTTCCGGCCGGNAAAGCGACACCCCAAGGATCCCGTGACA
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| MER52A | Spps | 117 | 127 | + | 19.34 | GCCCCGCCCCC |
| MER52A | PATZ1 | 117 | 127 | - | 19.16 | GGGGGCGGGGC |
| MER52A | NHLH2 | 237 | 252 | - | 18.96 | GGCTGCAGCTGCGCCC |
| MER52A | ZBED4 | 1497 | 1506 | + | 18.54 | CCCGCCCCGC |
| MER52A | Clamp | 1547 | 1560 | - | 18.51 | GAGCGAGCGAGCGC |
| MER52A | ZFP14 | 1012 | 1026 | - | 18.46 | GGAGACCCTGGAGTG |
| MER52A | ZNF281 | 118 | 127 | - | 18.37 | GGGGGCGGGG |
| MER52A | KLF16 | 117 | 127 | + | 18.24 | GCCCCGCCCCC |
| MER52A | SP3 | 117 | 127 | + | 18.17 | GCCCCGCCCCC |
| MER52A | KLF12 | 118 | 126 | - | 18.01 | GGGGCGGGG |
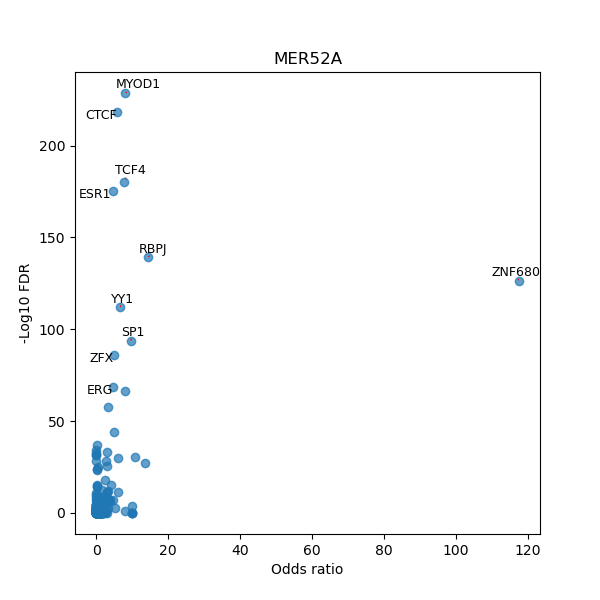
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.