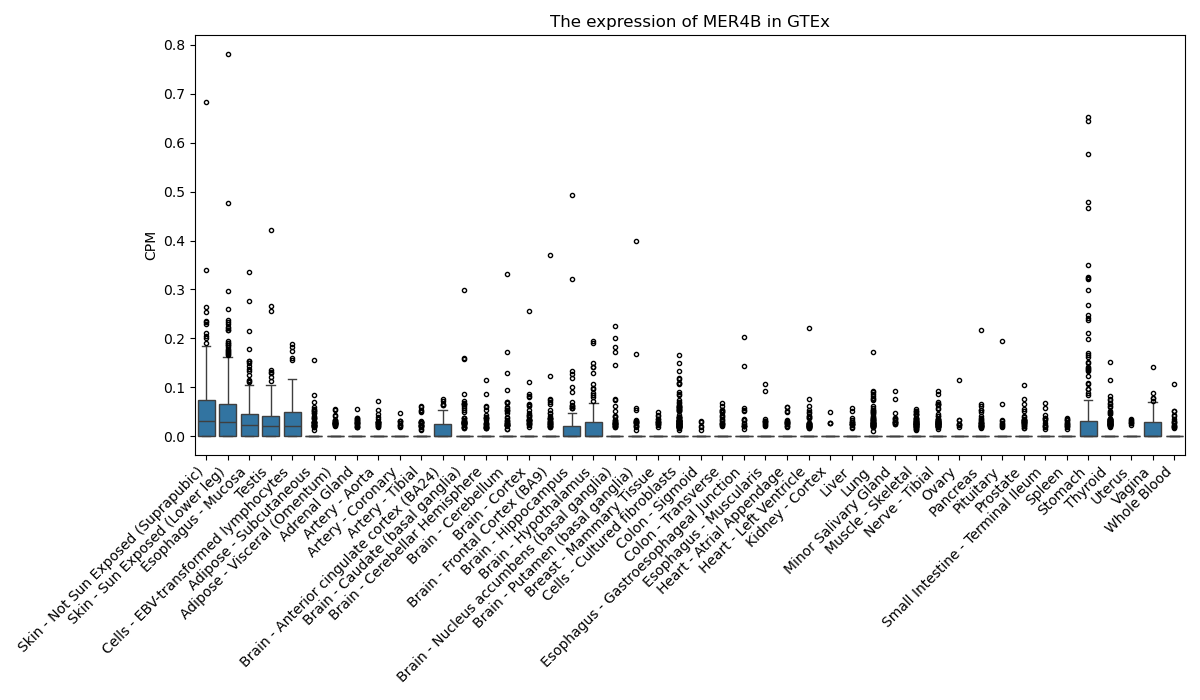
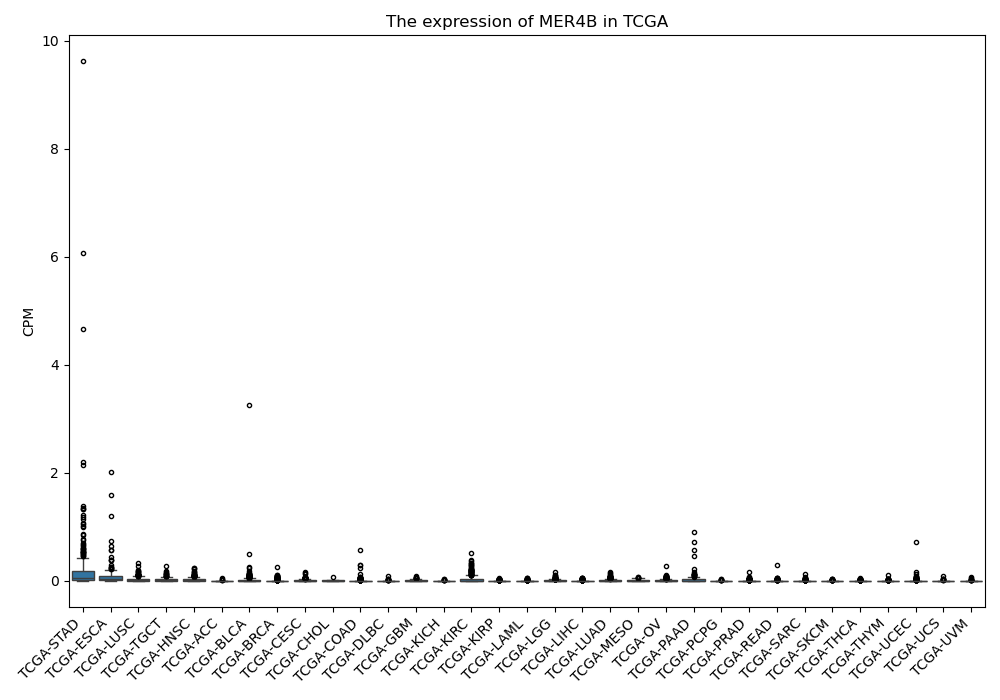
MER4B
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000797 |
|---|---|
| TE superfamily | ERV1 |
| TE class | LTR |
| Species | Haplorrhini |
| Length | 610 |
| Kimura value | 12.22 |
| Tau index | 0.9326 |
| Description | MER4B Long Terminal Repeat for ERV1 endogenous retrovirus |
| Comment | Note that MER stands for MEdium Reiteration frequency interspersed repeat. As such, MERs as a group are a grab bag of DNA transposons and retrotransposons. |
| Sequence |
TGTAAACCAAAAATAAAATTCTAAGCCCCCCAACCATCTGAATGGACCCCTCCTCTCGGCCAAGGGCATTCCAAAGTTAACCTGAAAAACTGGTTCAGGCCATGATGGGAAGNGGGGGTCGGACATGCCTCATTATACCCTCCTCCCTTTTGGAATTCAGGNANAGCCGACCAGCATTAACATCAACACAGACCTTAAGTCTGATAAGAAACATTTACAATCTATTCTCTCTGAAGCCTGCTACCTGGAGGCTTCATCTGCATGATAAAACCTTGGTCTCCACAACCCCTTATCGTAACCCAGACATTCCTTTCTATTGATAATAACTCTTTCAACCAATTGCCAATCAGAAAATCTTTAAATCTACCTATGACCTGGAAGCCCCCGCTTCGAGTTGTCCCGCCTTTCCGGACCGAACCAATGTACATCTTACATGTATTGATTGATGTCTCATGTCTCCCTAAAATGTATAAAACCAAGCTGTACCCCGACCACCTTGGGCACATGTTCTCAGGACCTCCTGAGGCTGTGTCACGGGCGCGTCCTTAACCTTGGCAAAATAAACTTTCTAAATTGATTGAGACCTGTCTCAGATACTTTTGGGTTCACA
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| MER4B | PBX1 | 439 | 447 | - | 15.97 | ATCAATCAA |
| MER4B | Pou5f1::Sox2 | 256 | 270 | - | 15.94 | TTTTATCATGCAGAT |
| MER4B | MEIS1 | 440 | 448 | - | 15.29 | CATCAATCA |
| MER4B | grh | 87 | 96 | - | 15.02 | GAACCAGTTT |
| MER4B | ceh-60 | 341 | 349 | - | 14.71 | TGATTGGCA |
| MER4B | cnd-1 | 33 | 40 | - | 14.66 | CAGATGGT |
| MER4B | NFYA | 343 | 350 | + | 14.54 | CCAATCAG |
| MER4B | elt-3 | 202 | 209 | - | 14.52 | TCTTATCA |
| MER4B | MEF2C | 7 | 17 | + | 14.47 | CCAAAAATAAA |
| MER4B | RIN | 7 | 16 | - | 14.47 | TTATTTTTGG |
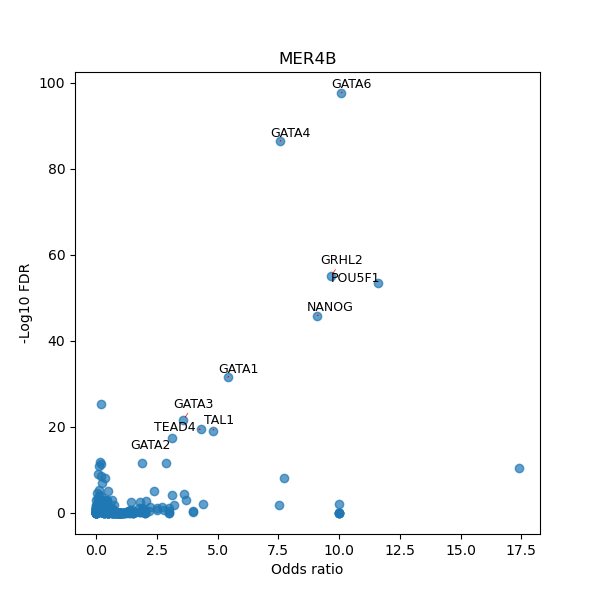
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.