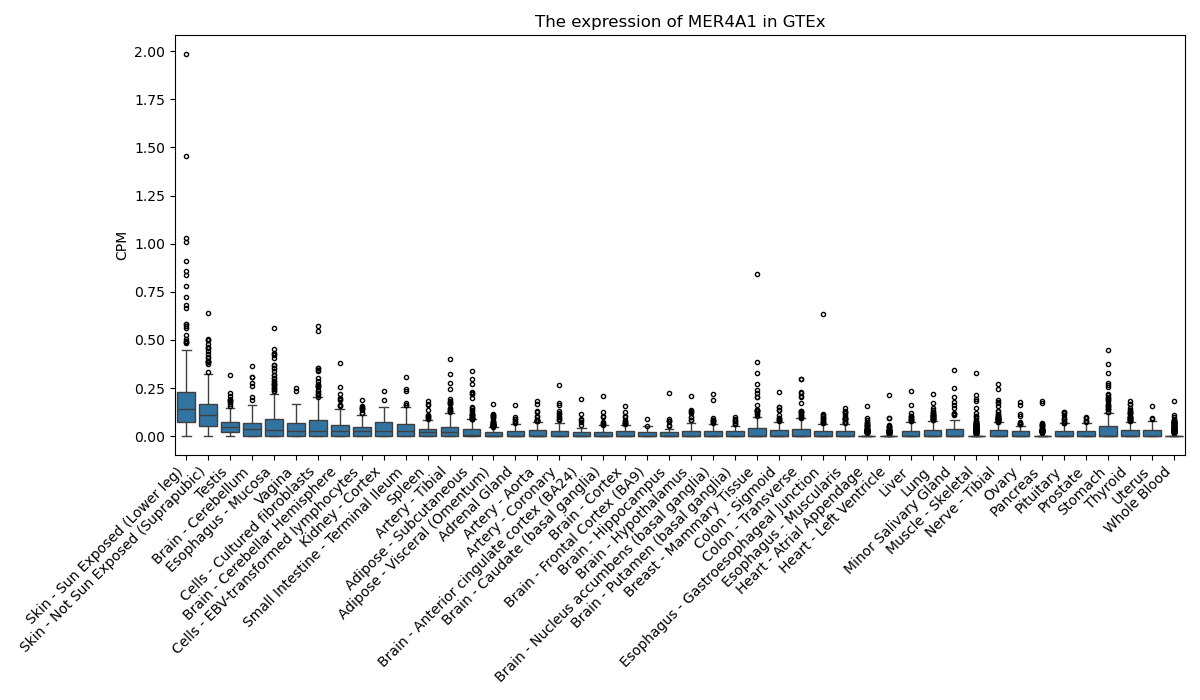
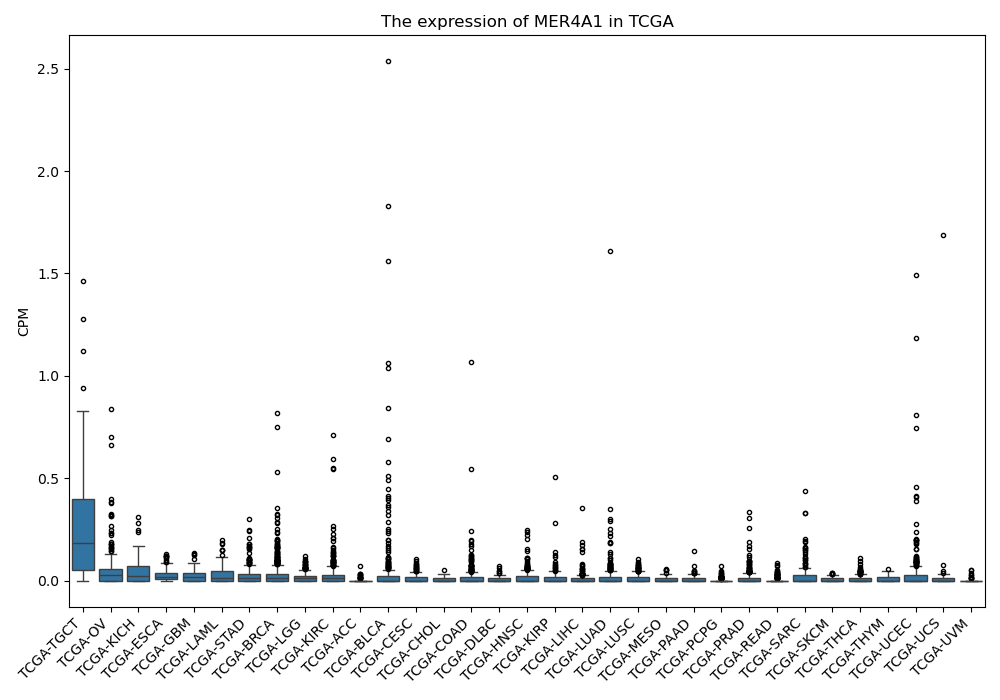
MER4A1
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000795 |
|---|---|
| TE superfamily | ERV1 |
| TE class | LTR |
| Species | Haplorrhini |
| Length | 472 |
| Kimura value | 9.11 |
| Tau index | 0.9306 |
| Description | MER4A1 Long Terminal Repeat for ERV1 endogenous retrovirus |
| Comment | Note that MER stands for MEdium Reiteration frequency interspersed repeat. As such, MERs as a group are a grab bag of DNA transposons and retrotransposons. |
| Sequence |
TGTGAAAGGAAAATAAATCTTGGGGCCCCAAAATCACTAAGCTAAAGGGAAAAGTCAAGCTGGGAACTGCTTAGGGCAAACCTGCCTCCCATTCTATTCAAAGTCACCCCTCTGCTCACTGAGATAAATGCATATCTGATTGCCTCCTTTGGAGAGGCTAATCAGAAACTCAAAAGAATGCAACCATTTGTCTCTCACCTACCTGTGACCTGGAAGCCCCCTCCCCGCTTCGAGTCNTCCCGCCTTTGCTTCGAGTTGTCCCGCCTTTCCGGACCGAACCAATGTNCATCTTACATATGTTGATTGATGTCTCATGTCTCCCTAAAATGTATAAAACCAAGCTGTGCCCCGACCACCTTGGGCACATGTCGTCAGGACCTCCTGAGGCTGTGTCACGGGCGCGCGTCCTCAACCTTGGCAAAATAAACTTTCTAAATTAACTGAGACCTGTCTCAGATTTTCGGGGTTCACA
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| MER4A1 | dsf | 50 | 57 | + | 14.52 | AAAAGTCA |
| MER4A1 | TBX19 | 193 | 209 | + | 14.48 | TCTCACCTACCTGTGAC |
| MER4A1 | GCR1 | 211 | 218 | + | 14.47 | TGGAAGCC |
| MER4A1 | CDF5 | 38 | 58 | - | 14.30 | TTGACTTTTCCCTTTAGCTTA |
| MER4A1 | SP2 | 218 | 226 | - | 14.29 | GGGGAGGGG |
| MER4A1 | tll | 50 | 59 | + | 14.27 | AAAAGTCAAG |
| MER4A1 | ZBTB7C | 350 | 357 | + | 14.24 | CGACCACC |
| MER4A1 | KLF5 | 217 | 226 | + | 14.20 | CCCCCTCCCC |
| MER4A1 | unc-86 | 128 | 135 | - | 14.03 | ATATGCAT |
| MER4A1 | KLF14 | 218 | 226 | - | 13.88 | GGGGAGGGG |
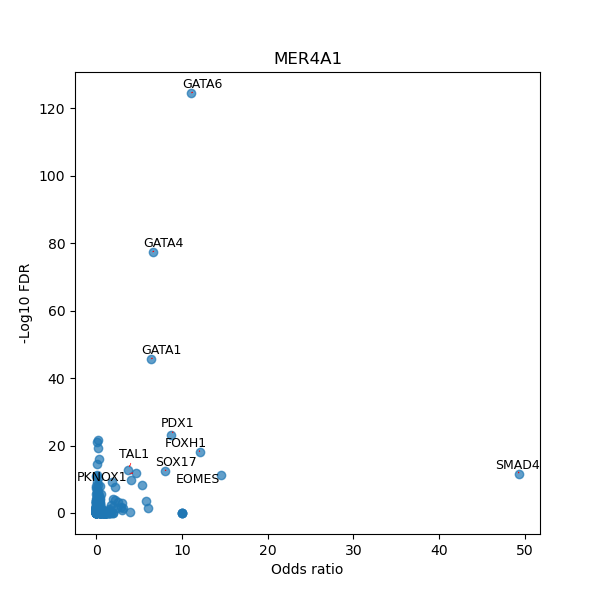
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.