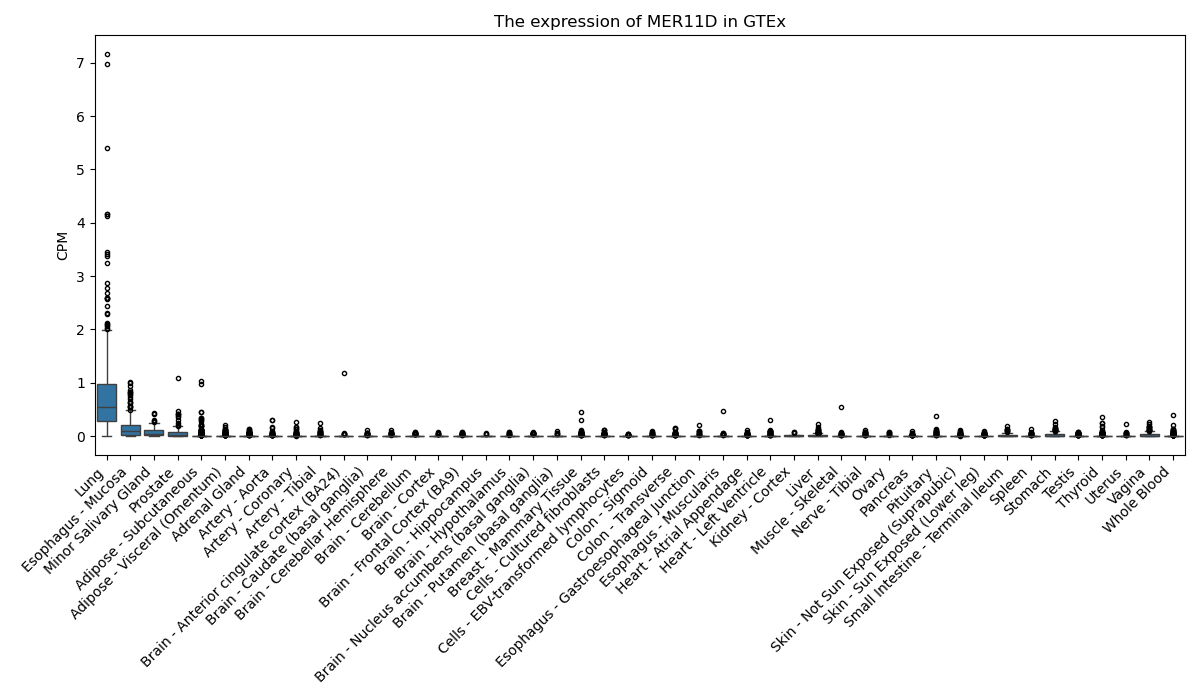
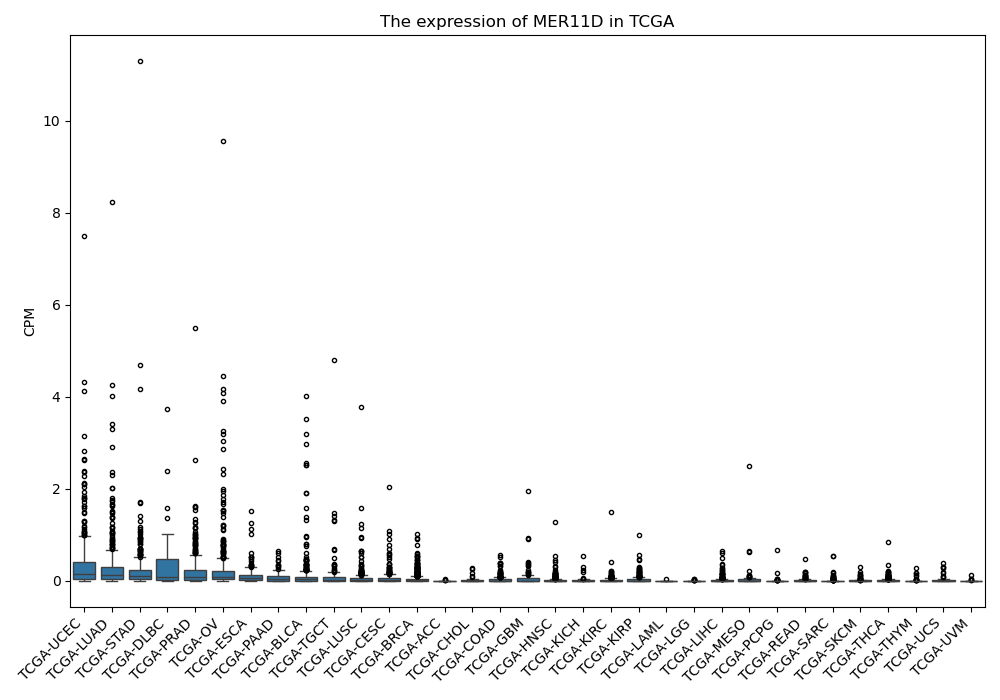
MER11D
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000718 |
|---|---|
| TE superfamily | ERV2 |
| TE class | LTR |
| Species | Catarrhini |
| Length | 897 |
| Kimura value | 3.28 |
| Tau index | 0.9929 |
| Description | MER11D Long Terminal Repeat for HERVK11 endogenous retrovirus |
| Comment | MER11D is a retroviral LTR that has been proliferated by the HERVK- related retrovirus HERVK11. Generates 6 bp TSDs. MER11D is a very young subfamily of MER11 LTRs, 98% similarity to the consensus sequence. MER11D consensus sequence is only 64% similar to MER11A,B,C consensus sequences, which are also young subfamilies (~90% similarity). Note that MER stands for MEdium Reiteration frequency interspersed repeat. As such, MERs as a group are a grab bag of DNA transposons and retrotransposons. |
| Sequence |
TGTAGGGAGACCCCCTGAAACTATTGCTACGGAATAAAAGATGAAATGCTCCTGATTATTGTAAATACAAAATTGCATGCAGGATTGTGTAAAGACAATGCCAGGTTGGACTGCCAGAACGAGCCAACAGCGCGTGATGTGCTTCCCCCTGCAGAGAGCCTATGAATGGACGTGCAGTCAGGGAGGTTTCACATCACCAAGATTCCTATCCCAGAAAAGCAGATGTTCATAGCTCTGGGAATGGAATGCGACCCTTGTGGAGAGCCTATAAACGGACGCATGGGGGGCGCCTGTCCATATGGATAAGATAGGGCTATAAACGCCCTCATCTTGCCACGGCTCTTCTAGGCCTCTTTAGGGTTAAGGCATACTCCCTTCTGAGAATTTCTGGTCTAACCGGTTGTCTAGCTTCACGTCCTGTTTCTATGGATTGTTTGTAACCAGCTTTTGCTGCAACTGTTACTGCTGATTAATATCTTGCTAATCATAGGTTATGGAAAGACTGTGTTTCTGTTTTAAGGCTCTGTTAGAAATTACTGATGCACACACTATATTGTAAATTCTTATCTCTGTATACTGTACTTCTGCATACAGATGTACTGTACTTCTACATACAAATGTTATGTTAAAGAATTACTTCATCCCCATGTGACCATCTCACCTCATAATCAAATGACCCTAAATCCCTCACTAACCTACCCCCGCCCTCACTAAACTTAATAATAAATGCTGGTATATCCAGTGCATTGTTGGCACCGCGGGACCAGAAGGCGGTGACCCCCCTGGACCCAGCTTTCACTATCTTGTGTGTGTCTATTATTTCTCGACCTGCCGATCCGCCTGGGAACAAAGAGAGAGCCCCGTTGCATTGCGGGCTGCTGGCCAGATCCCGCAATA
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| MER11D | AT5G56840 | 297 | 310 | - | 17.74 | TATCTTATCCATAT |
| MER11D | AT1G19000 | 297 | 309 | + | 17.41 | ATATGGATAAGAT |
| MER11D | AT5G61620 | 297 | 310 | + | 17.33 | ATATGGATAAGATA |
| MER11D | KLF15 | 700 | 707 | + | 16.27 | CCCCGCCC |
| MER11D | cg | 805 | 815 | - | 16.11 | AGACACACACA |
| MER11D | ZNF76 | 860 | 876 | - | 16.03 | GCCCGCAATGCAACGGG |
| MER11D | USV1 | 10 | 19 | + | 15.98 | ACCCCCTGAA |
| MER11D | PATZ1 | 699 | 709 | - | 15.97 | GAGGGCGGGGG |
| MER11D | KLF7 | 700 | 707 | - | 15.72 | GGGCGGGG |
| MER11D | Stat2 | 508 | 517 | - | 15.65 | AAAACAGAAA |
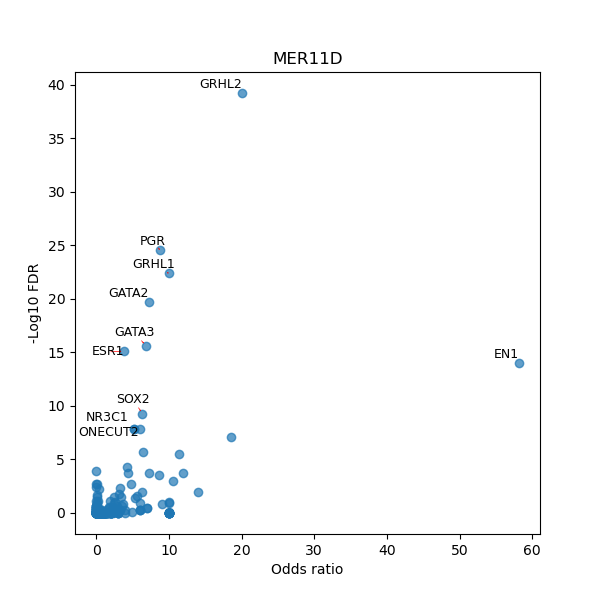
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.