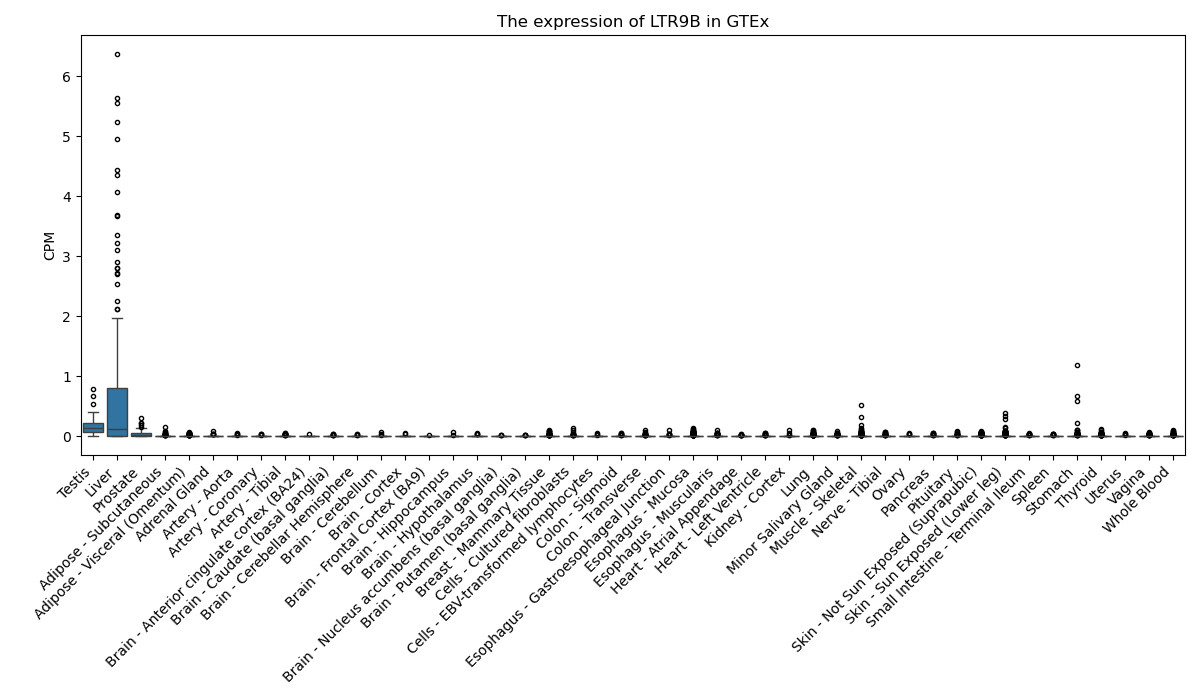
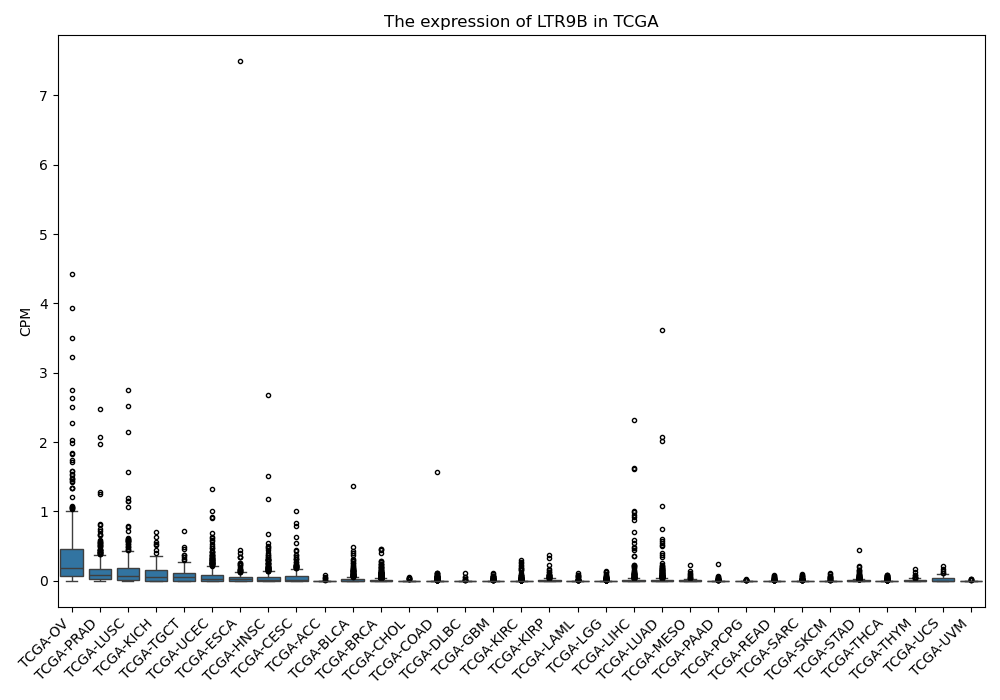
LTR9B
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000623 |
|---|---|
| TE superfamily | ERV1 |
| TE class | LTR |
| Species | Haplorrhini |
| Length | 644 |
| Kimura value | 9.68 |
| Tau index | 0.9765 |
| Description | LTR9B (Long Terminal Repeat) for ERV1 endogenous retrovirus |
| Comment | LTR9B is similar to LTR9 over its full length and is found flanking closely related internal sequences (mostly HUERS-P3b). 4 bp TSDs. HUERS-P3 LTR. |
| Sequence |
TGTTGTAGGAGTTATTAAGAAATTATTTTAGGCAGATAGAGAGGAAAAGGGGTCCTTGGGAAGTTTTCGTTTCTTTTTAAAGCAGCTCCAGAAACGTTTCTTGTCTAGCAGGAAAGCCCCGGCTCTTAGAGCCGGGCCGGCAACCTTTGATATGCAAATGCAGGCCATTAGAAACTGGGTCCACCCAACATGGCGATTCCCGCCGTCGTCTTCTTGCCCTTGCCCCACATGTGCCTGGCAACATGGCCGCCCCCACATATCCCCACGTGTGTAGAACATCATGGCGCCCTGCATTTGCATATTAAAAGGCTAGGGTGGGAGGGCCAGTTTTTTCGCGGGCTACGTGAATGACATGCCTGGTCAAACCAATCCCCTGAGCCCTATGCAAATCAGACACCGCCTCCTCCAGCCTCCTCATATAACTGGCTGGTNTCCGCCGCACNCGGGGTTTCCTCTCTCGGCTTTGGAGCCCCCCTCCCTCTGTCTCTGTACGGGGGAGCTTCTTCCTTCTTTCTTCTCCCTTCTTTCTTGCCTATTAAACTCTCCGCTCCTTAAAACCACTCCACGTGTGTCCGTGTCGTTTTATCTAANTCGGCGCGAGGACCAAGAACCCTGGTGTTCCTCCACTCATCGGAGCCGTATCA
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| LTR9B | BPC1 | 27 | 50 | + | -0.71 | TTTAGGCAGATAGAGAGGAAAAGG |
| LTR9B | EWSR1-FLI1 | 502 | 519 | - | -1.04 | GAGAAGAAAGAAGGAAGA |
| LTR9B | BPC1 | 503 | 526 | - | -1.23 | AAGAAGGGAGAAGAAAGAAGGAAG |
| LTR9B | BPC1 | 466 | 489 | - | -1.50 | CAGAGACAGAGGGAGGGGGGCTCC |
| LTR9B | BPC1 | 499 | 522 | - | -1.68 | AGGGAGAAGAAAGAAGGAAGAAGC |
| LTR9B | BPC1 | 505 | 528 | - | -2.00 | GAAAGAAGGGAGAAGAAAGAAGGA |
| LTR9B | EWSR1-FLI1 | 516 | 533 | - | -12.42 | GGCAAGAAAGAAGGGAGA |
| LTR9B | EWSR1-FLI1 | 512 | 529 | - | -16.13 | AGAAAGAAGGGAGAAGAA |
| LTR9B | DAL81 | 196 | 214 | - | -26.29 | AGAAGACGACGGCGGGAAT |
| LTR9B | DAL81 | 304 | 322 | + | -39.42 | AAAAGGCTAGGGTGGGAGG |
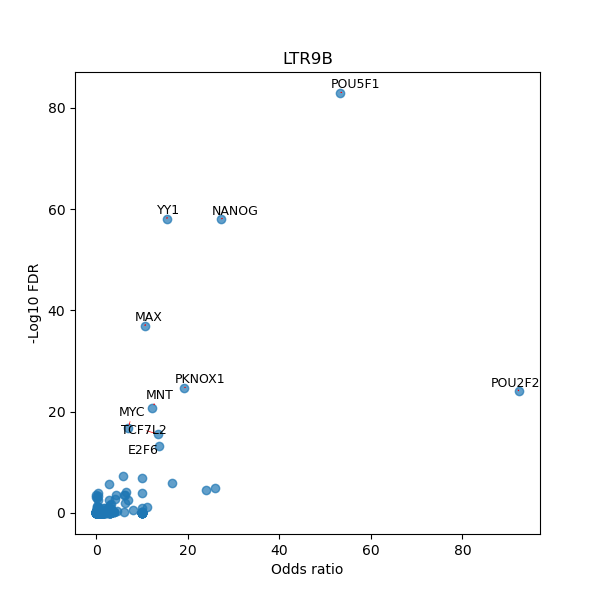
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.