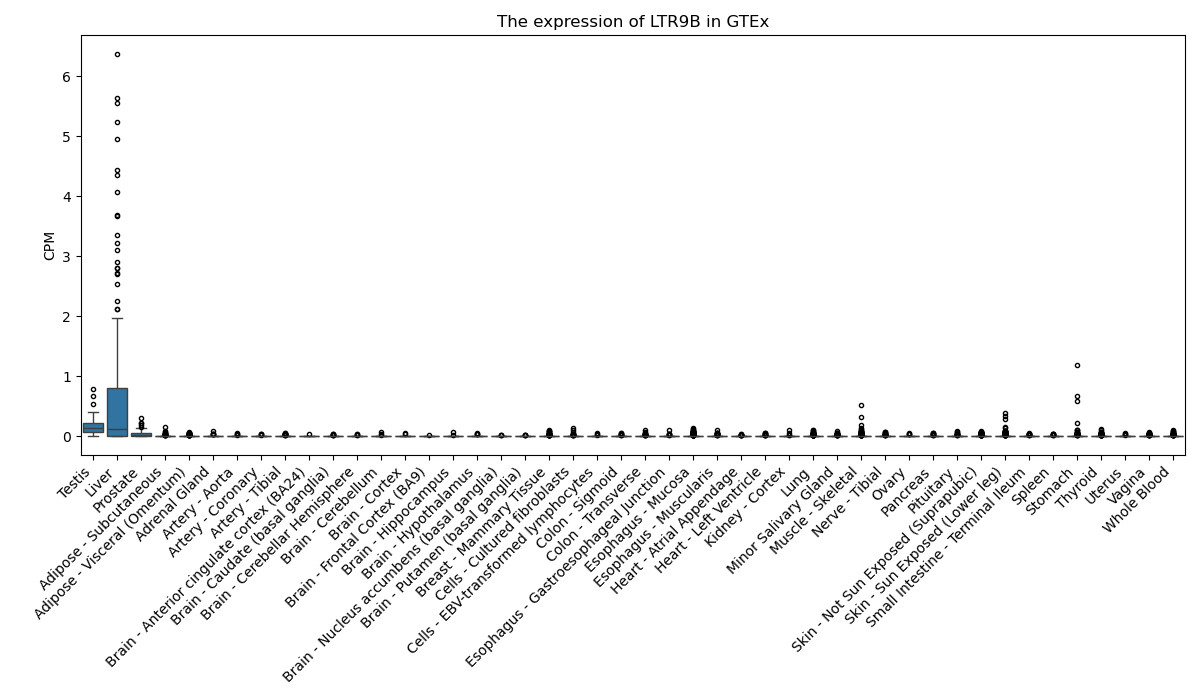
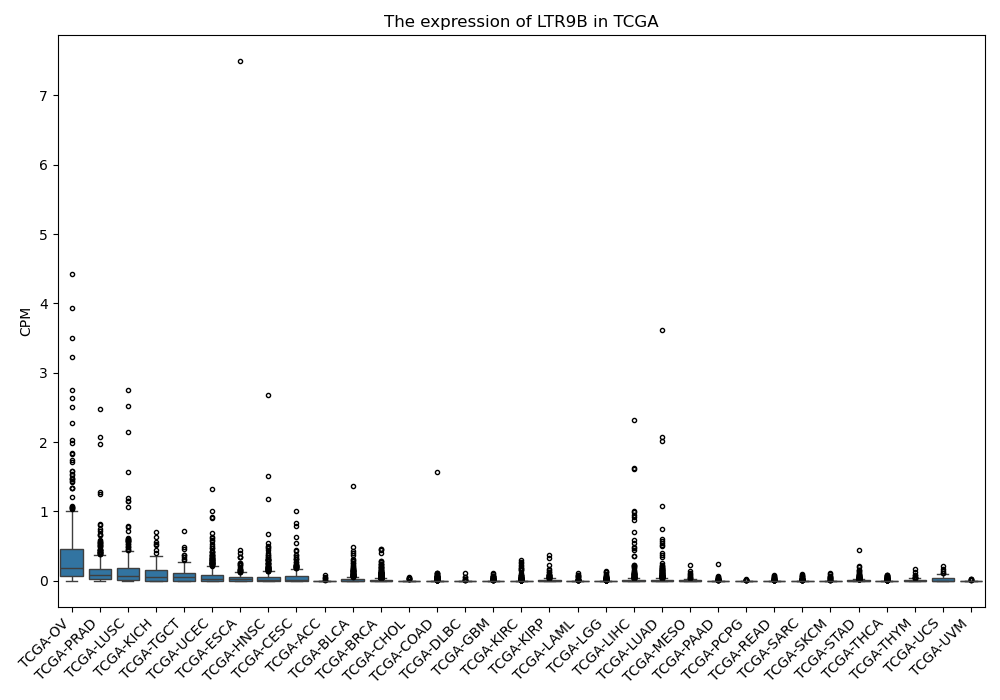
LTR9B
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000623 |
|---|---|
| TE superfamily | ERV1 |
| TE class | LTR |
| Species | Haplorrhini |
| Length | 644 |
| Kimura value | 9.68 |
| Tau index | 0.9765 |
| Description | LTR9B (Long Terminal Repeat) for ERV1 endogenous retrovirus |
| Comment | LTR9B is similar to LTR9 over its full length and is found flanking closely related internal sequences (mostly HUERS-P3b). 4 bp TSDs. HUERS-P3 LTR. |
| Sequence |
TGTTGTAGGAGTTATTAAGAAATTATTTTAGGCAGATAGAGAGGAAAAGGGGTCCTTGGGAAGTTTTCGTTTCTTTTTAAAGCAGCTCCAGAAACGTTTCTTGTCTAGCAGGAAAGCCCCGGCTCTTAGAGCCGGGCCGGCAACCTTTGATATGCAAATGCAGGCCATTAGAAACTGGGTCCACCCAACATGGCGATTCCCGCCGTCGTCTTCTTGCCCTTGCCCCACATGTGCCTGGCAACATGGCCGCCCCCACATATCCCCACGTGTGTAGAACATCATGGCGCCCTGCATTTGCATATTAAAAGGCTAGGGTGGGAGGGCCAGTTTTTTCGCGGGCTACGTGAATGACATGCCTGGTCAAACCAATCCCCTGAGCCCTATGCAAATCAGACACCGCCTCCTCCAGCCTCCTCATATAACTGGCTGGTNTCCGCCGCACNCGGGGTTTCCTCTCTCGGCTTTGGAGCCCCCCTCCCTCTGTCTCTGTACGGGGGAGCTTCTTCCTTCTTTCTTCTCCCTTCTTTCTTGCCTATTAAACTCTCCGCTCCTTAAAACCACTCCACGTGTGTCCGTGTCGTTTTATCTAANTCGGCGCGAGGACCAAGAACCCTGGTGTTCCTCCACTCATCGGAGCCGTATCA
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| LTR9B | Pou5f1::Sox2 | 145 | 159 | + | 20.97 | CTTTGATATGCAAAT |
| LTR9B | POU2F1::SOX2 | 144 | 160 | - | 19.90 | CATTTGCATATCAAAGG |
| LTR9B | ZFP42 | 239 | 251 | + | 16.53 | CAACATGGCCGCC |
| LTR9B | nub | 382 | 392 | + | 16.52 | TATGCAAATCA |
| LTR9B | Pou5f1::Sox2 | 293 | 307 | - | 16.48 | TTTTAATATGCAAAT |
| LTR9B | POU2F1::SOX2 | 292 | 308 | + | 16.24 | CATTTGCATATTAAAAG |
| LTR9B | MAZ | 472 | 479 | + | 16.08 | CCCCTCCC |
| LTR9B | ZNF148 | 472 | 481 | + | 16.08 | CCCCTCCCTC |
| LTR9B | Zfp961 | 282 | 289 | - | 15.99 | GGGCGCCA |
| LTR9B | POU2F2 | 150 | 162 | + | 15.88 | ATATGCAAATGCA |
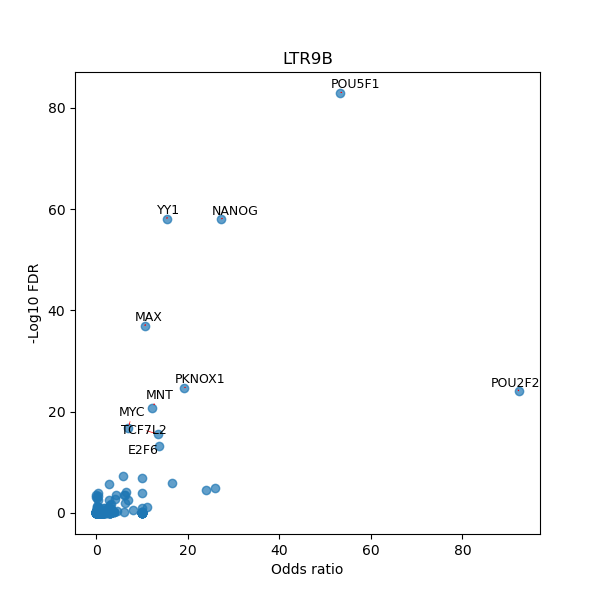
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.