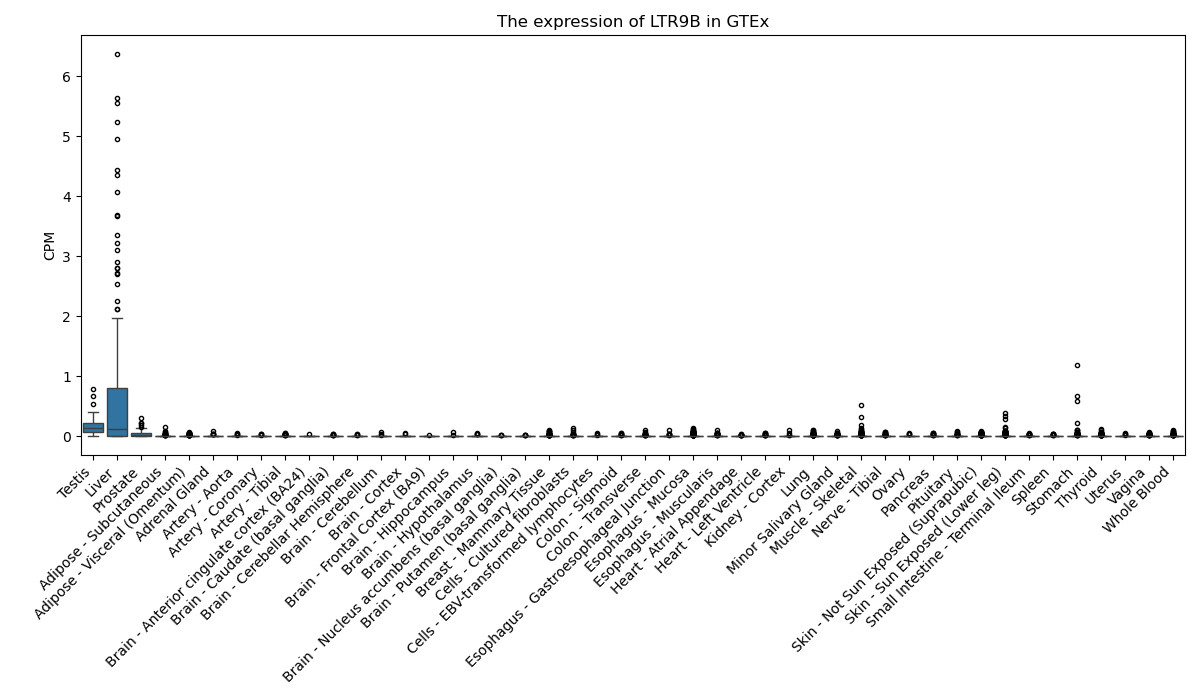
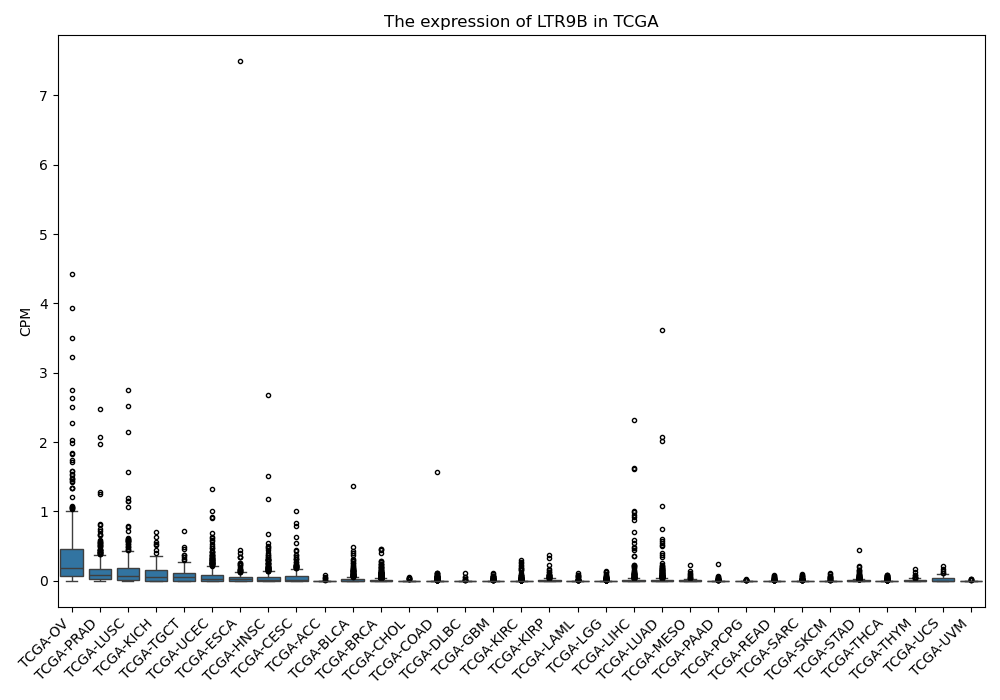
LTR9B
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000623 |
|---|---|
| TE superfamily | ERV1 |
| TE class | LTR |
| Species | Haplorrhini |
| Length | 644 |
| Kimura value | 9.68 |
| Tau index | 0.9765 |
| Description | LTR9B (Long Terminal Repeat) for ERV1 endogenous retrovirus |
| Comment | LTR9B is similar to LTR9 over its full length and is found flanking closely related internal sequences (mostly HUERS-P3b). 4 bp TSDs. HUERS-P3 LTR. |
| Sequence |
TGTTGTAGGAGTTATTAAGAAATTATTTTAGGCAGATAGAGAGGAAAAGGGGTCCTTGGGAAGTTTTCGTTTCTTTTTAAAGCAGCTCCAGAAACGTTTCTTGTCTAGCAGGAAAGCCCCGGCTCTTAGAGCCGGGCCGGCAACCTTTGATATGCAAATGCAGGCCATTAGAAACTGGGTCCACCCAACATGGCGATTCCCGCCGTCGTCTTCTTGCCCTTGCCCCACATGTGCCTGGCAACATGGCCGCCCCCACATATCCCCACGTGTGTAGAACATCATGGCGCCCTGCATTTGCATATTAAAAGGCTAGGGTGGGAGGGCCAGTTTTTTCGCGGGCTACGTGAATGACATGCCTGGTCAAACCAATCCCCTGAGCCCTATGCAAATCAGACACCGCCTCCTCCAGCCTCCTCATATAACTGGCTGGTNTCCGCCGCACNCGGGGTTTCCTCTCTCGGCTTTGGAGCCCCCCTCCCTCTGTCTCTGTACGGGGGAGCTTCTTCCTTCTTTCTTCTCCCTTCTTTCTTGCCTATTAAACTCTCCGCTCCTTAAAACCACTCCACGTGTGTCCGTGTCGTTTTATCTAANTCGGCGCGAGGACCAAGAACCCTGGTGTTCCTCCACTCATCGGAGCCGTATCA
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| LTR9B | nub | 151 | 161 | + | 15.55 | TATGCAAATGC |
| LTR9B | nub | 291 | 301 | - | 15.55 | TATGCAAATGC |
| LTR9B | RELA | 111 | 120 | - | 15.52 | GGGGCTTTCC |
| LTR9B | POU1F1 | 290 | 303 | + | 15.51 | TGCATTTGCATATT |
| LTR9B | TCP16 | 177 | 184 | - | 15.32 | GTGGACCC |
| LTR9B | POU2F2 | 381 | 393 | + | 15.31 | CTATGCAAATCAG |
| LTR9B | E2F6 | 197 | 204 | - | 15.29 | GGCGGGAA |
| LTR9B | PATZ1 | 471 | 481 | - | 15.22 | GAGGGAGGGGG |
| LTR9B | REL | 111 | 120 | - | 15.13 | GGGGCTTTCC |
| LTR9B | Wt1 | 247 | 256 | + | 15.11 | CCGCCCCCAC |
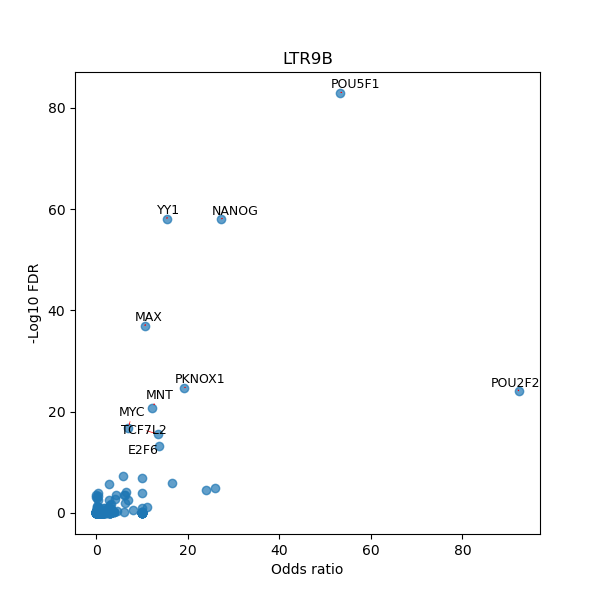
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.