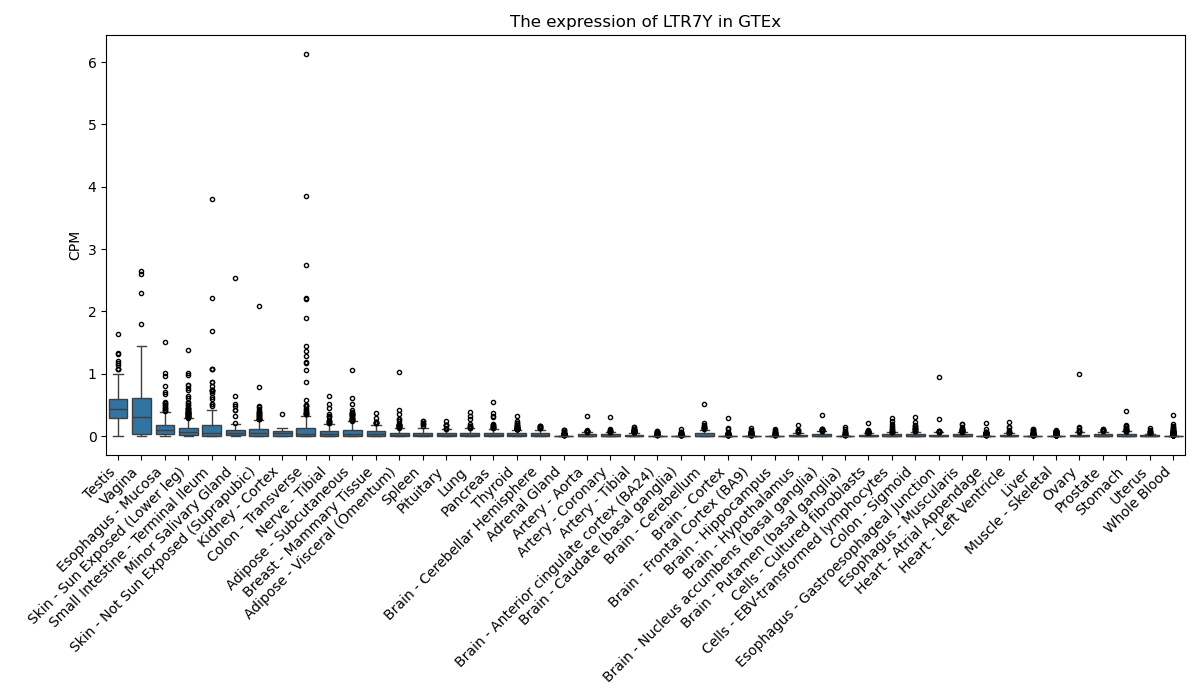
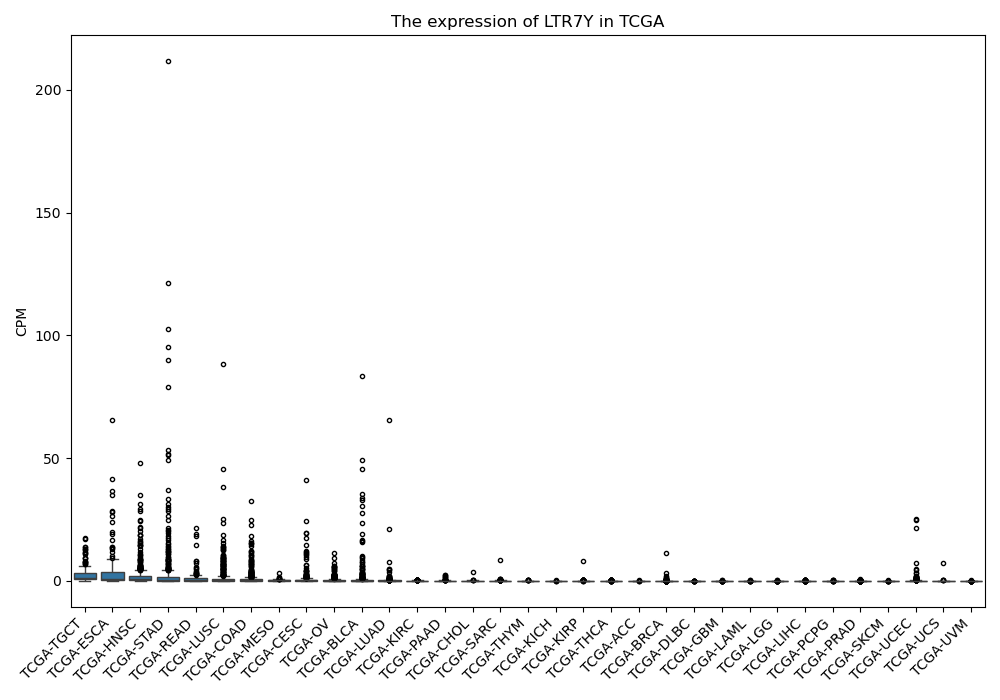
LTR7Y
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000589 |
|---|---|
| TE superfamily | ERV1 |
| TE class | LTR |
| Species | Hominidae |
| Length | 464 |
| Kimura value | 5.96 |
| Tau index | 0.9508 |
| Description | Long terminal repeat of an HERVH endogenous retrovirus |
| Comment | LTR of an HERVH element active 7-18 MYA, mostly in the common ancestor of gorilla, human and chimp. Updated from 2005 entry. Two copies are in the human hg38 genome are precisely absent from the chimpanzee genome: a solo LTR at chr11:49842787-49843262 with surprisingly many mutations, and a copy with a partial internal sequence at chr9:86586832-86589058. A 21-bp sequence starting at pos 174 is present in 1 or 2 tandem copies. |
| Sequence |
TGTCAGGCCTCTGAGCCCAGGCCAGGCCATCGCATCCCCTGTGACTTGCACGTATACATCCAGATGGCCTGAAGTAACTGAAGATCCACAAAAGAAGTAAAAACAGCCTTAACTGATGACATTCCACCATTGTGATTTGTTCCTGCCCCACCCTAACTGATCAATGTACTTTGTAATCTCCCCCACCCTTGCTTTGTAGTCTCCCCCACCCTTAAGAAGGTTCTTTGTAATTCTCCCCACCCTTGAGAATGTACTTTGTGAGATCCACCCCTGCCCACCAGAGAACAACCCCCTTTGACTGTAATTTTCCATTACCTTCCCAAATCCTATAAAACGGCCCCACCCCTATCTCCCTTCGCTGACTCTCTTTTCGGACTCAGCCCGCCTGCACCCAGGTGAAATAAACAGCCATGTTGCTCACACAAAGCCTGTTTGGTGGTCTCTTCACACGGACGCGCATGAAA
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| LTR7Y | KLF5 | 234 | 243 | + | 14.12 | TCCCCACCCT |
| LTR7Y | nhr-86 | 194 | 201 | + | 14.10 | TTGTAGTC |
| LTR7Y | ZNF213 | 15 | 26 | + | 14.06 | GCCCAGGCCAGG |
| LTR7Y | KLF5 | 180 | 189 | + | 14.06 | CCCCCACCCT |
| LTR7Y | KLF5 | 203 | 212 | + | 14.06 | CCCCCACCCT |
| LTR7Y | LjSGA_053525.1 | 336 | 343 | + | 14.06 | GGCCCCAC |
| LTR7Y | Nfatc2 | 306 | 313 | - | 14.03 | AATGGAAA |
| LTR7Y | MYB10 | 430 | 439 | - | 13.90 | CCACCAAACA |
| LTR7Y | KLF16 | 337 | 347 | + | 13.84 | GCCCCACCCCT |
| LTR7Y | KLF14 | 338 | 346 | - | 13.80 | GGGGTGGGG |
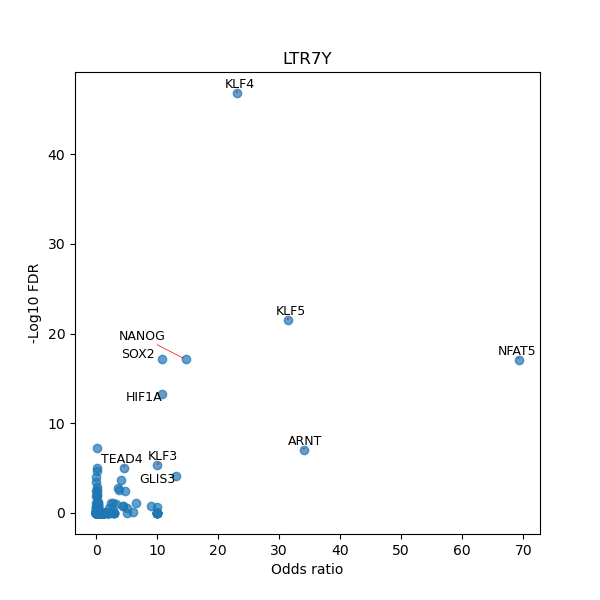
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.