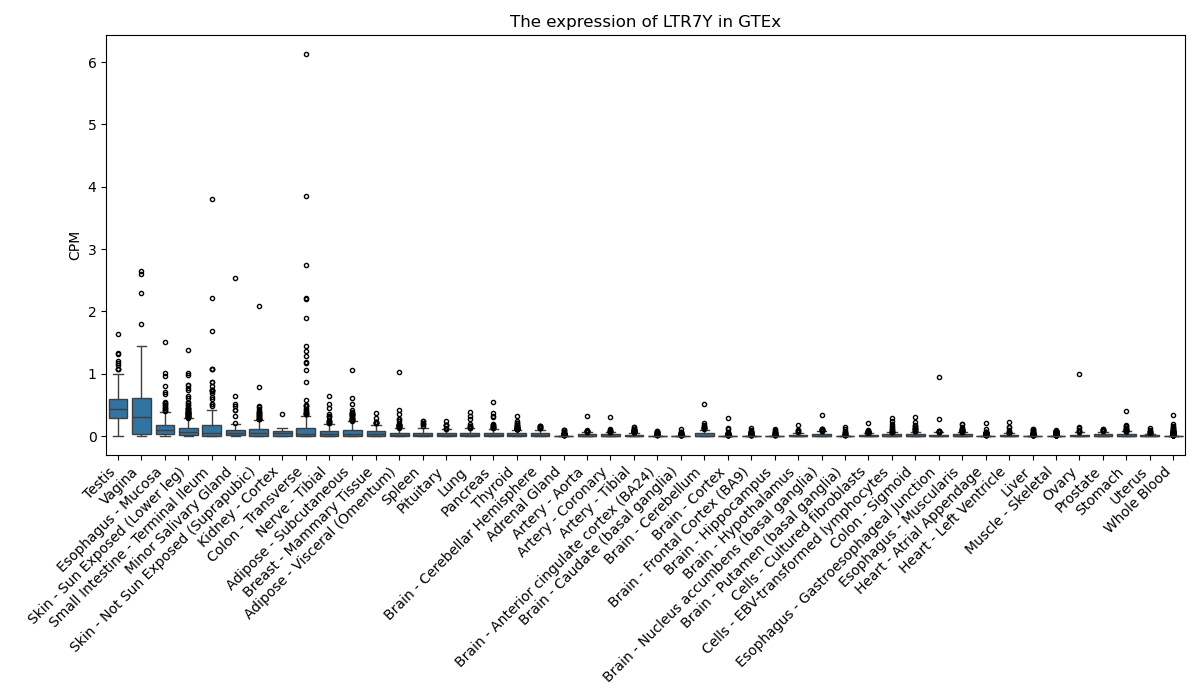
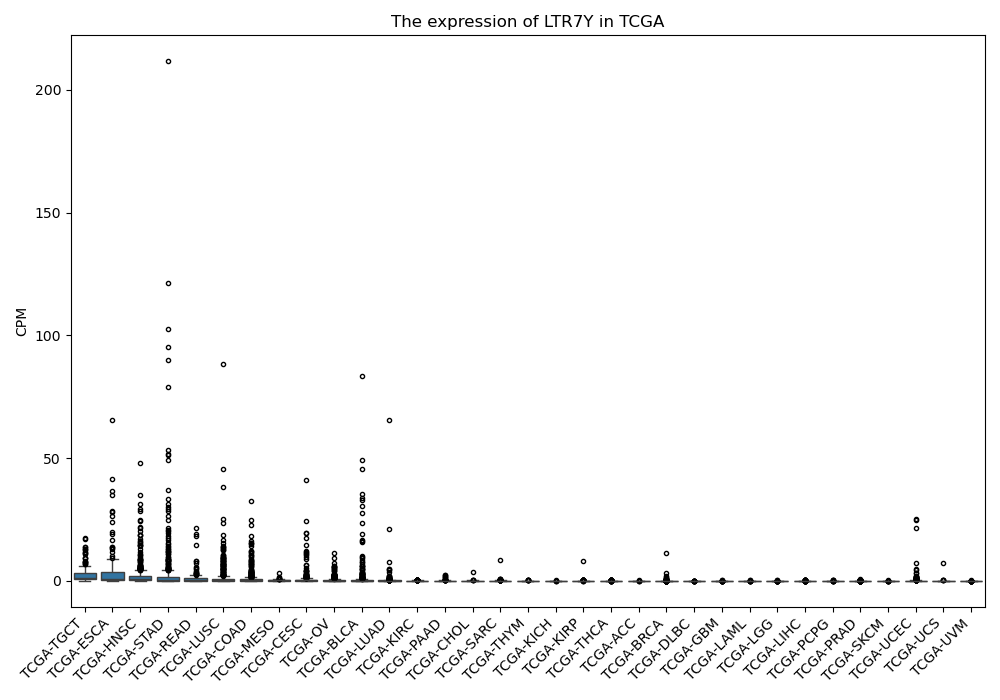
LTR7Y
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000589 |
|---|---|
| TE superfamily | ERV1 |
| TE class | LTR |
| Species | Hominidae |
| Length | 464 |
| Kimura value | 5.96 |
| Tau index | 0.9508 |
| Description | Long terminal repeat of an HERVH endogenous retrovirus |
| Comment | LTR of an HERVH element active 7-18 MYA, mostly in the common ancestor of gorilla, human and chimp. Updated from 2005 entry. Two copies are in the human hg38 genome are precisely absent from the chimpanzee genome: a solo LTR at chr11:49842787-49843262 with surprisingly many mutations, and a copy with a partial internal sequence at chr9:86586832-86589058. A 21-bp sequence starting at pos 174 is present in 1 or 2 tandem copies. |
| Sequence |
TGTCAGGCCTCTGAGCCCAGGCCAGGCCATCGCATCCCCTGTGACTTGCACGTATACATCCAGATGGCCTGAAGTAACTGAAGATCCACAAAAGAAGTAAAAACAGCCTTAACTGATGACATTCCACCATTGTGATTTGTTCCTGCCCCACCCTAACTGATCAATGTACTTTGTAATCTCCCCCACCCTTGCTTTGTAGTCTCCCCCACCCTTAAGAAGGTTCTTTGTAATTCTCCCCACCCTTGAGAATGTACTTTGTGAGATCCACCCCTGCCCACCAGAGAACAACCCCCTTTGACTGTAATTTTCCATTACCTTCCCAAATCCTATAAAACGGCCCCACCCCTATCTCCCTTCGCTGACTCTCTTTTCGGACTCAGCCCGCCTGCACCCAGGTGAAATAAACAGCCATGTTGCTCACACAAAGCCTGTTTGGTGGTCTCTTCACACGGACGCGCATGAAA
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| LTR7Y | KLF5 | 337 | 346 | + | 16.67 | GCCCCACCCC |
| LTR7Y | HSFB2A | 215 | 224 | + | 16.11 | AGAAGGTTCT |
| LTR7Y | KLF5 | 145 | 154 | + | 15.96 | GCCCCACCCT |
| LTR7Y | HSFB2B | 215 | 225 | + | 15.78 | AGAAGGTTCTT |
| LTR7Y | Wt1 | 177 | 186 | + | 15.69 | TCTCCCCCAC |
| LTR7Y | Wt1 | 200 | 209 | + | 15.69 | TCTCCCCCAC |
| LTR7Y | KLF4 | 146 | 153 | + | 15.48 | CCCCACCC |
| LTR7Y | KLF4 | 181 | 188 | + | 15.48 | CCCCACCC |
| LTR7Y | KLF4 | 204 | 211 | + | 15.48 | CCCCACCC |
| LTR7Y | KLF4 | 235 | 242 | + | 15.48 | CCCCACCC |
TFBS enrichment in GRCh38
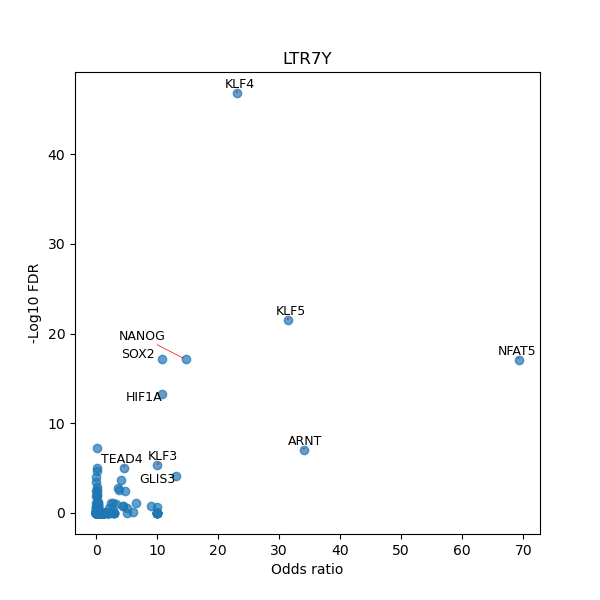
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.