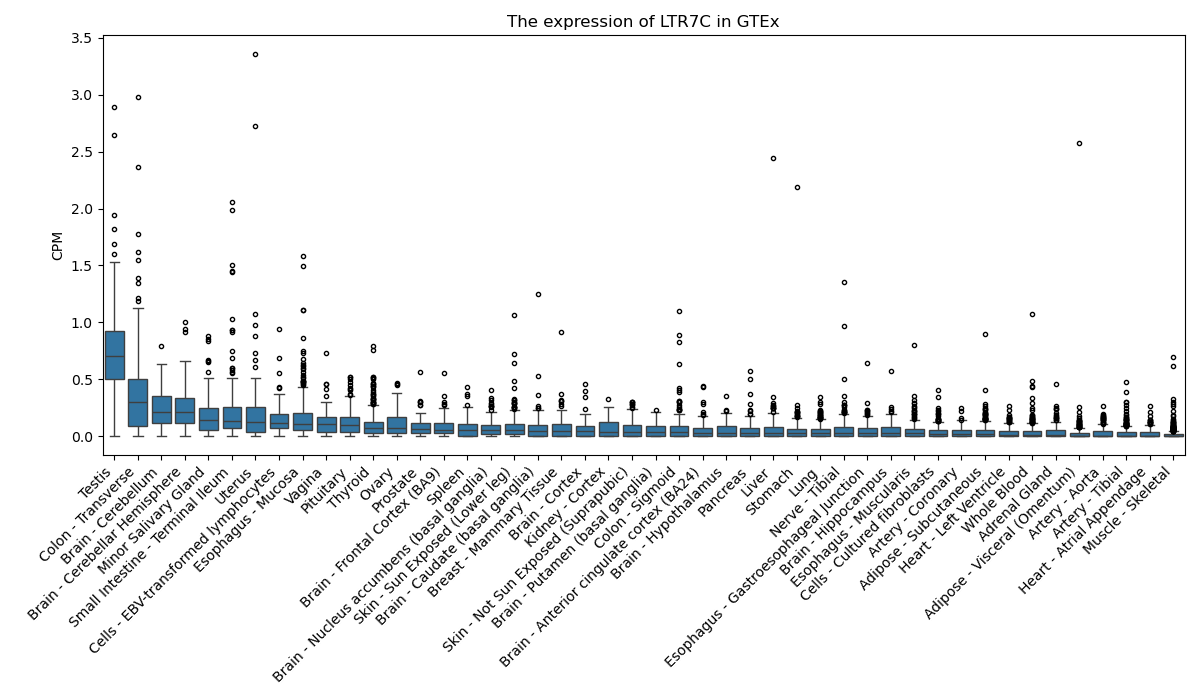
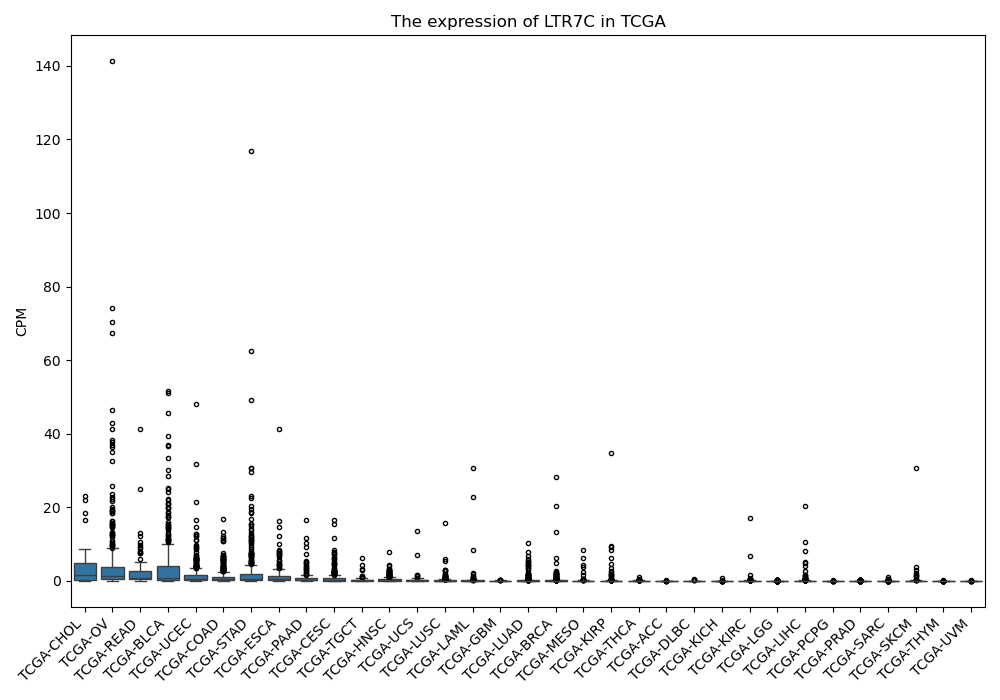
LTR7C
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000588 |
|---|---|
| TE superfamily | ERV1 |
| TE class | LTR |
| Species | Catarrhini |
| Length | 470 |
| Kimura value | 10.47 |
| Tau index | 0.9171 |
| Description | Long terminal repeat of an HERVH endogenous retrovirus |
| Comment | LTR of an HERVH element active 25-40 MYA. At least 3 distinct subfamilies in this set, but each with very few copies. |
| Sequence |
TGTCAGGCCTCTGAGCCGAAGCTCAGCCATTGTAACCCCTGTGACCTGCACATATACGTCCAGATGGCCTGCAGGAGCCAAGAAGTCTGGAGCAGCCGAAAAACCACAAAAGAAGTGAAACAGCCAGTTCCTGCCTTAACTGATTAACCAACCTTACGACATTCCACCATTGTGACTTGTCCCTGCCCTACCCCAACTGATCAATCGACCTTGTGACATTCTTCTTCTGGACAATGAGTCTTATGATCTCCCCACCATGCACCTTGTGACCCCCTCCTCTGCTGACAATAGATAACCACCTTTNACTGTAACTTTCCACTGCCTACCCAAGTCCTATAAAGCNGCCCCTCCCCTATCTCCCTTCGCTGACTCTCTTTTCGGACTCAGCCCACTTGCACCCAAGTGAATAAACAGCCTTGTTGCTCACACAAAGCCTGTTTGGTGGTCTCTTCACACGGACGCGCTTGACA
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| LTR7C | MYB107 | 437 | 444 | - | 14.64 | CACCAAAC |
| LTR7C | usp | 265 | 273 | - | 14.63 | GGGGTCACA |
| LTR7C | TEAD4 | 159 | 166 | + | 14.45 | ACATTCCA |
| LTR7C | bHLH130 | 389 | 396 | + | 14.38 | CCACTTGC |
| LTR7C | SP2 | 345 | 353 | - | 14.29 | GGGGAGGGG |
| LTR7C | PATZ1 | 270 | 280 | - | 14.25 | AGAGGAGGGGG |
| LTR7C | MAZ | 271 | 278 | + | 14.06 | CCCCTCCT |
| LTR7C | RARA::RXRA | 173 | 189 | - | 14.00 | AGGGCAGGGACAAGTCA |
| LTR7C | bHLH112 | 389 | 395 | + | 13.92 | CCACTTG |
| LTR7C | OsFBH1 | 389 | 395 | + | 13.92 | CCACTTG |
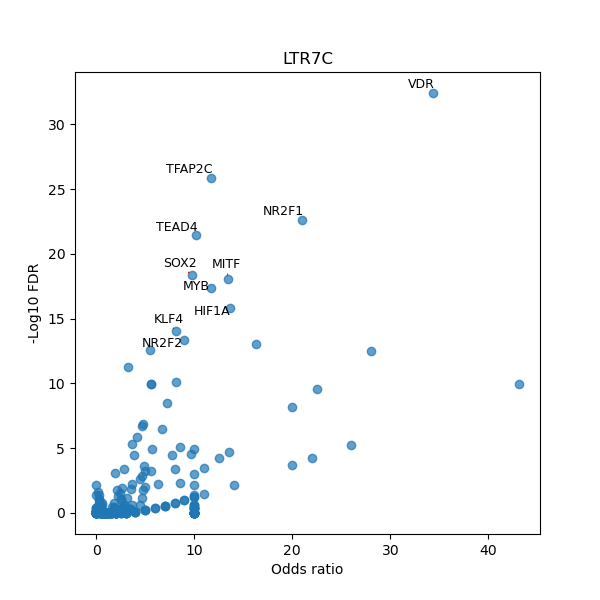
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.