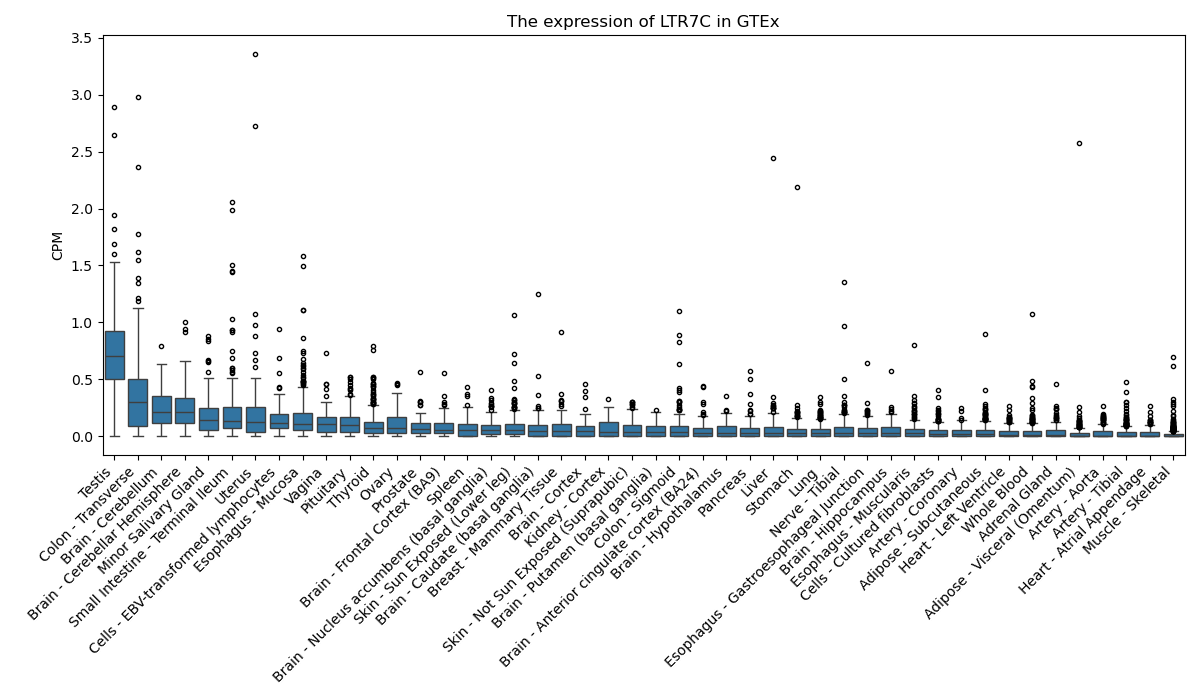
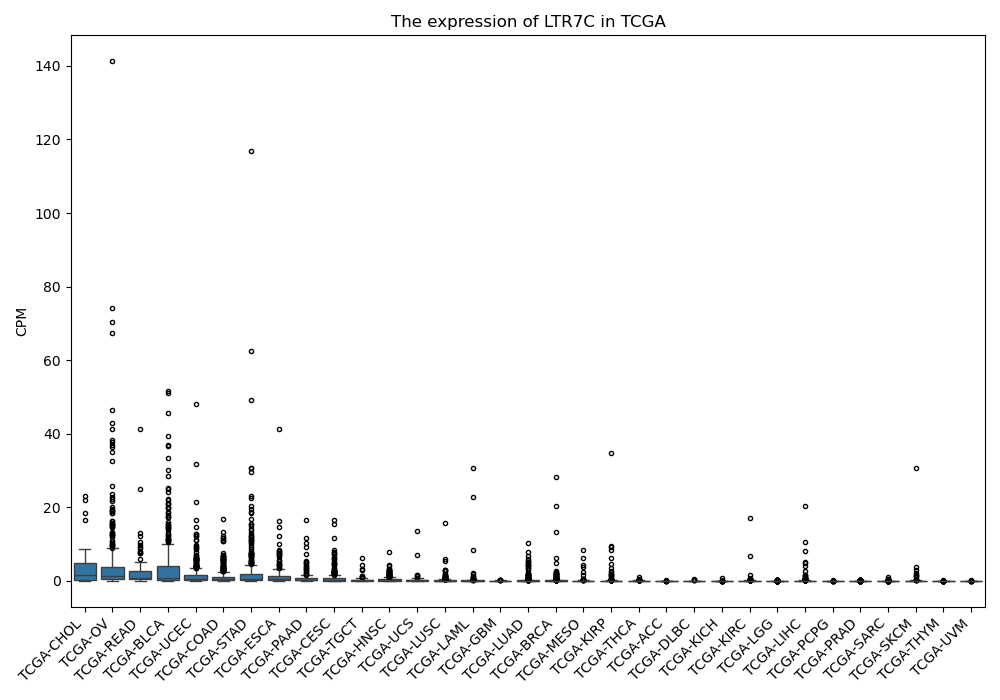
LTR7C
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000588 |
|---|---|
| TE superfamily | ERV1 |
| TE class | LTR |
| Species | Catarrhini |
| Length | 470 |
| Kimura value | 10.47 |
| Tau index | 0.9171 |
| Description | Long terminal repeat of an HERVH endogenous retrovirus |
| Comment | LTR of an HERVH element active 25-40 MYA. At least 3 distinct subfamilies in this set, but each with very few copies. |
| Sequence |
TGTCAGGCCTCTGAGCCGAAGCTCAGCCATTGTAACCCCTGTGACCTGCACATATACGTCCAGATGGCCTGCAGGAGCCAAGAAGTCTGGAGCAGCCGAAAAACCACAAAAGAAGTGAAACAGCCAGTTCCTGCCTTAACTGATTAACCAACCTTACGACATTCCACCATTGTGACTTGTCCCTGCCCTACCCCAACTGATCAATCGACCTTGTGACATTCTTCTTCTGGACAATGAGTCTTATGATCTCCCCACCATGCACCTTGTGACCCCCTCCTCTGCTGACAATAGATAACCACCTTTNACTGTAACTTTCCACTGCCTACCCAAGTCCTATAAAGCNGCCCCTCCCCTATCTCCCTTCGCTGACTCTCTTTTCGGACTCAGCCCACTTGCACCCAAGTGAATAAACAGCCTTGTTGCTCACACAAAGCCTGTTTGGTGGTCTCTTCACACGGACGCGCTTGACA
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| LTR7C | ZNF76 | 250 | 266 | + | 16.41 | CCCCACCATGCACCTTG |
| LTR7C | KLF5 | 344 | 353 | + | 16.10 | GCCCCTCCCC |
| LTR7C | ZNF148 | 345 | 354 | + | 16.10 | CCCCTCCCCT |
| LTR7C | MAZ | 345 | 352 | + | 16.08 | CCCCTCCC |
| LTR7C | SP1 | 345 | 353 | - | 16.06 | GGGGAGGGG |
| LTR7C | PATZ1 | 344 | 354 | - | 16.01 | AGGGGAGGGGC |
| LTR7C | SP4 | 345 | 353 | - | 15.85 | GGGGAGGGG |
| LTR7C | bHLH80 | 389 | 397 | - | 15.52 | TGCAAGTGG |
| LTR7C | GATA1::TAL1 | 279 | 295 | - | 15.15 | TTATCTATTGTCAGCAG |
| LTR7C | Bcl11B | 101 | 109 | + | 15.12 | AAACCACAA |
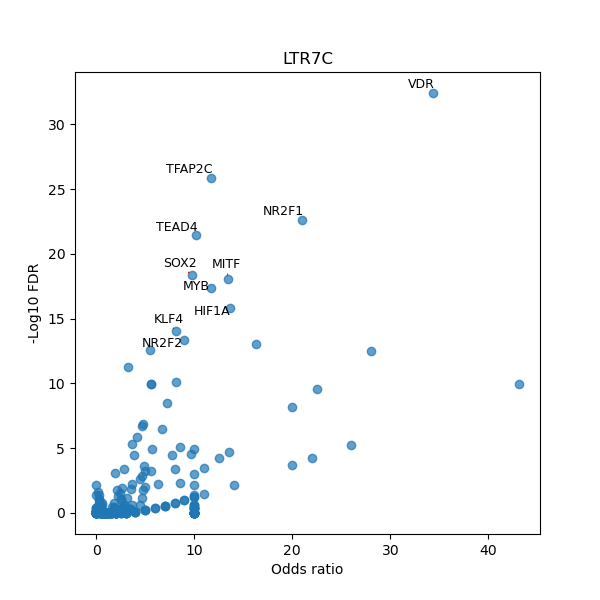
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.