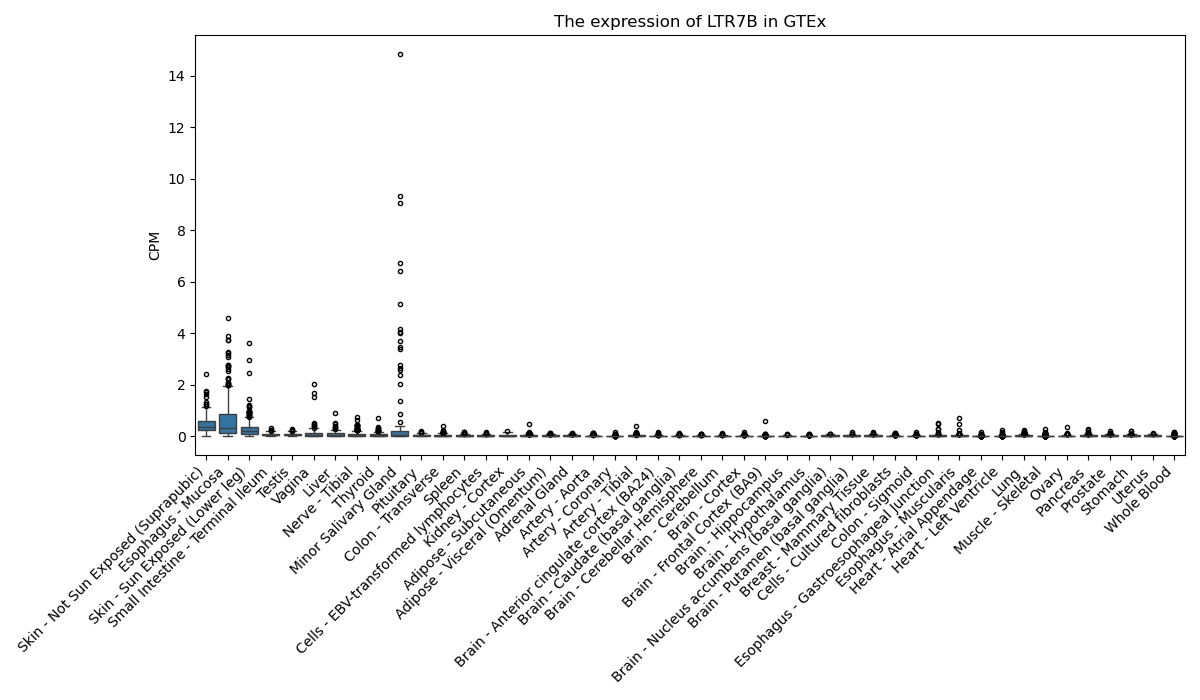
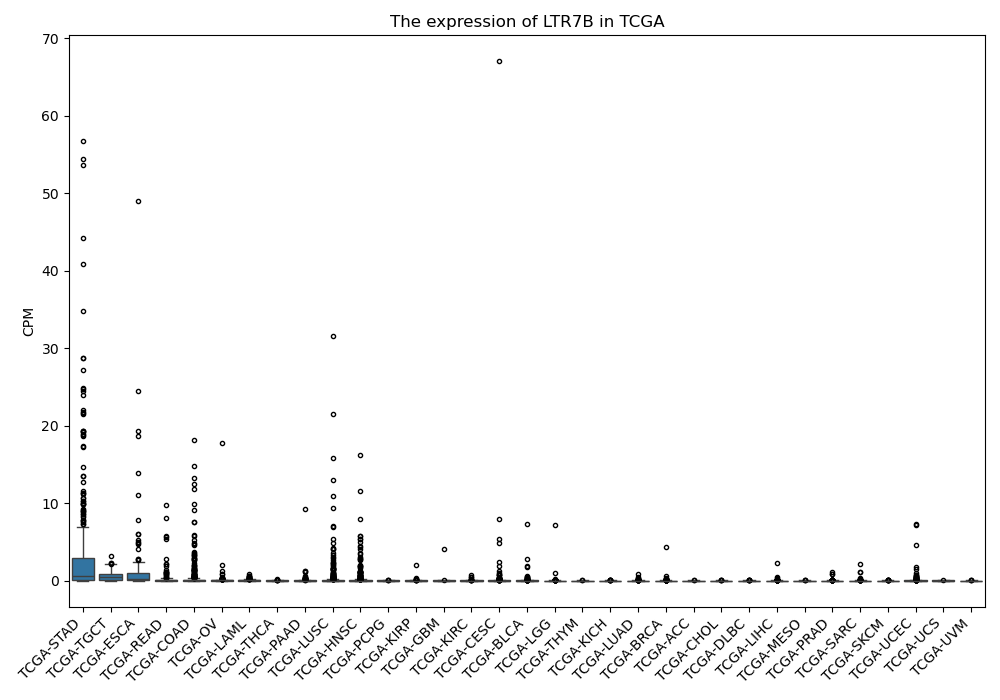
LTR7B
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000587 |
|---|---|
| TE superfamily | ERV1 |
| TE class | LTR |
| Species | Catarrhini |
| Length | 464 |
| Kimura value | 4.82 |
| Tau index | 0.9462 |
| Description | Long terminal repeat of an HERVH endogenous retrovirus |
| Comment | LTR of an HERVH element active 15 to 40 MYA. About half the copies predate our speciation from Old World monkeys, about 10% is unique to the great ape genomes. A 1 bp sequence starting around pos 180 appears with 1 or 2 copies. The version with 1 copy was named LTR7B1 in the reference. |
| Sequence |
TGTCAGGCCTCTGAGCCCAAGCTAAGCCATCATATCCCCTGTGACCTGCACGTATACATCCAGATGGCCTGAAGCAACTGAAGATCCACAAAAGAAGTGAAAATAGCCTTAACTGATGACATTCCACCATTGTGATTTGTTTCTGCCCCACCCTAACTGATCAATGTACTTTGTAATCTCCCCCACCCTTAAGAAGGTTCTTTGTAATCTCCCCCACCCTTAAGAAGGTTCTTTGTAATTCTCCCCACCCTTGAGAATGTACTTTGTGAGATCCACCCCCTGCCCGCAAAACATTGCTCCTAACTCCACCGCCTATCCCAAAACCTATAAGAACTAATGATAATCCCACCACCCTTTGCTGACTCTCTTTTCGGACTCAGCCCGCCTGCACCCAGGTGAAATAAACAGCCTTGTTGCTCACACAAAGCCTGTTTGGTGGTCTCTTCACACGGACGCGTGAGACA
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| LTR7B | KLF5 | 180 | 189 | + | 14.06 | CCCCCACCCT |
| LTR7B | KLF5 | 211 | 220 | + | 14.06 | CCCCCACCCT |
| LTR7B | TBXT | 445 | 460 | - | 14.04 | TCACGCGTCCGTGTGA |
| LTR7B | MYB10 | 430 | 439 | - | 13.90 | CCACCAAACA |
| LTR7B | TBXT | 445 | 460 | + | 13.88 | TCACACGGACGCGTGA |
| LTR7B | ERF054 | 306 | 313 | - | 13.80 | GGCGGTGG |
| LTR7B | dar1 | 145 | 155 | + | 13.64 | GCCCCACCCTA |
| LTR7B | MYB13 | 430 | 439 | - | 13.60 | CCACCAAACA |
| LTR7B | NEUROD1 | 60 | 67 | + | 13.36 | CCAGATGG |
| LTR7B | MYB58 | 431 | 439 | - | 13.35 | CCACCAAAC |
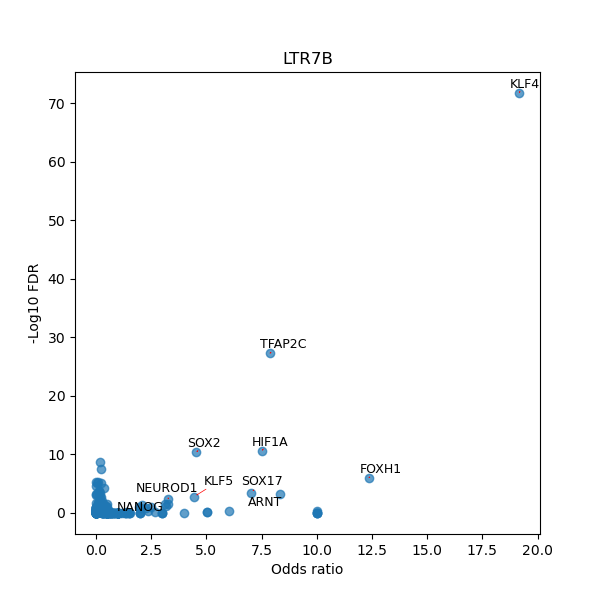
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.