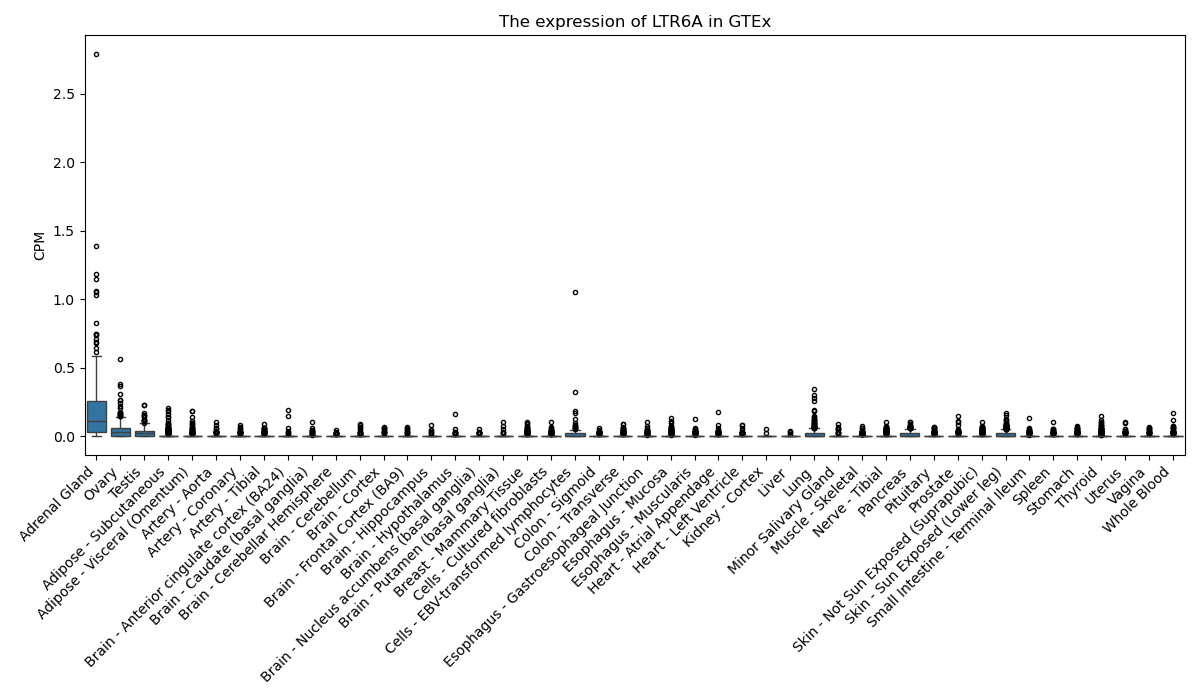
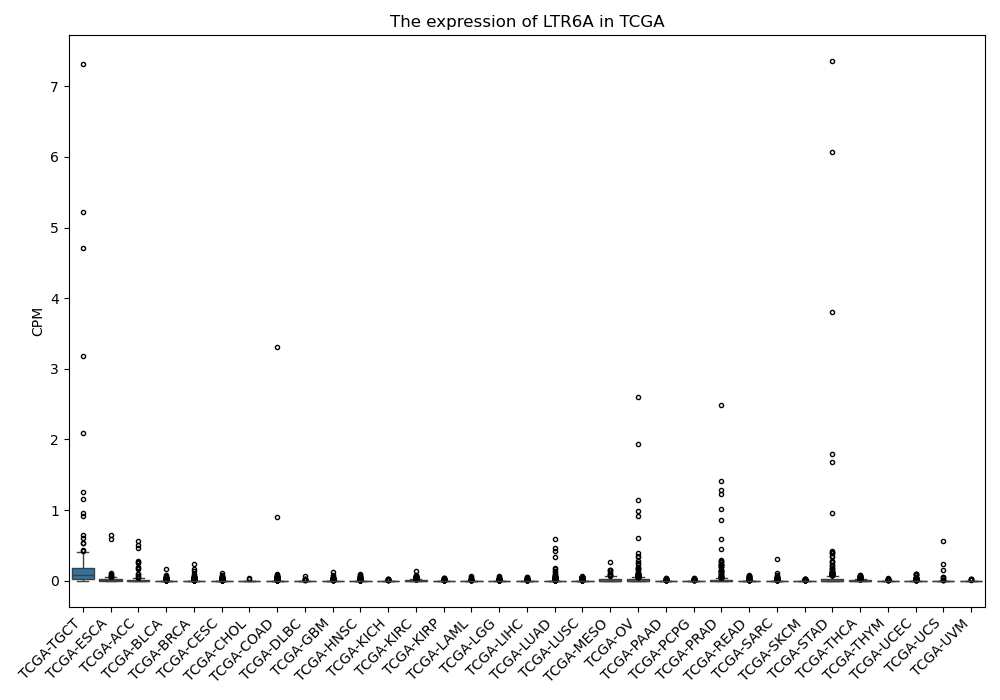
LTR6A
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000569 |
|---|---|
| TE superfamily | ERV1 |
| TE class | LTR |
| Species | Catarrhini |
| Length | 564 |
| Kimura value | 14.78 |
| Tau index | 0.9907 |
| Description | LTR6A (Long Terminal Repeat) for HERVS71 endogenous retrovirus |
| Comment | Some copies present at orthologous site in old world monkeys, others specific to apes. |
| Sequence |
TGTGATACCCTACCTTGTTTTAACCTGAATNGACTCTCCCTTAGCTGAGAGAGCCGGACGGACTCCATTTGGCTCCTTCACTTGCAAGACATCAAGGGCTCCTTACCCACCCCCTTCCTCAAGGACTTAACTTGTGCAAGCTGACTCCCAGCACATCAAAGAGTGCAATTAACTGATAAGGTACTGNGGCAAGCTATGTCCGCAGTTCCCAGGAATTCGCCCGGNTGATAGTACCCTAAAGCCCCCGCGTTTGTGTCCGGTNGATAGCACCCAAAGCCCCCGCACCTATCACCTTGTGATGGATTTAAAGCCCCTGCACCTGGAACTGTTTGTTTTCCTGTAACCATTTGTCTTTTTAACTTTTTTGCCTGTTTTGCTTCTGTAAGATTGCTTCAGCTAGGCTCCCCCTCCCCTTTCTAAACCAAAGTATAAAAGAAAATCTAGCCCCTTCTTCGGGGCCGAGAGAATTTTGAGCGCTAGCCGTCTCTCGGTCGCCGGCTAATAAAGGACTCCTGAATTCGTCTCAAAGTGTGGCGTTTCTCTATAACTCGCTCGGTTACAACA
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| LTR6A | DOF5.8 | 347 | 365 | + | 18.85 | TTTGTCTTTTTAACTTTTT |
| LTR6A | Msgn1 | 343 | 352 | - | 17.20 | GACAAATGGT |
| LTR6A | DOF3.6 | 419 | 439 | - | 16.67 | TTTTCTTTTATACTTTGGTTT |
| LTR6A | JKD | 346 | 357 | + | 16.61 | ATTTGTCTTTTT |
| LTR6A | DOF3.4 | 349 | 365 | + | 16.47 | TGTCTTTTTAACTTTTT |
| LTR6A | PATZ1 | 404 | 414 | - | 16.43 | AGGGGAGGGGG |
| LTR6A | ZNF530 | 404 | 417 | - | 16.32 | GAAAGGGGAGGGGG |
| LTR6A | IDD11 | 346 | 357 | - | 16.31 | AAAAAGACAAAT |
| LTR6A | DOF3.6 | 348 | 368 | + | 16.15 | TTGTCTTTTTAACTTTTTTGC |
| LTR6A | ZNF148 | 405 | 414 | + | 16.10 | CCCCTCCCCT |
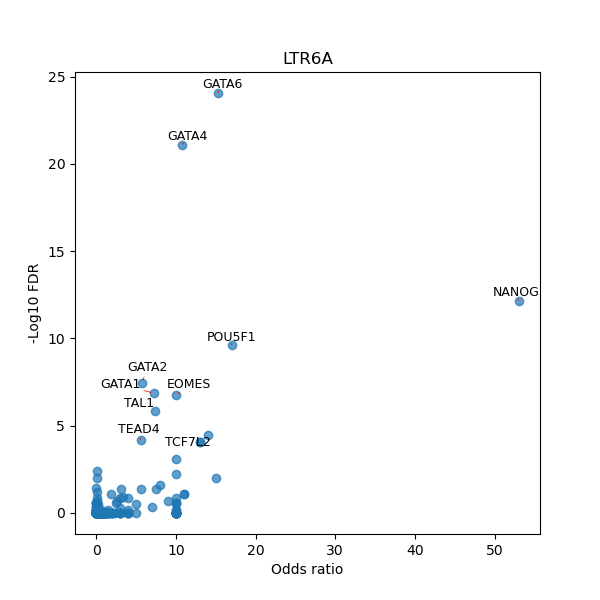
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.