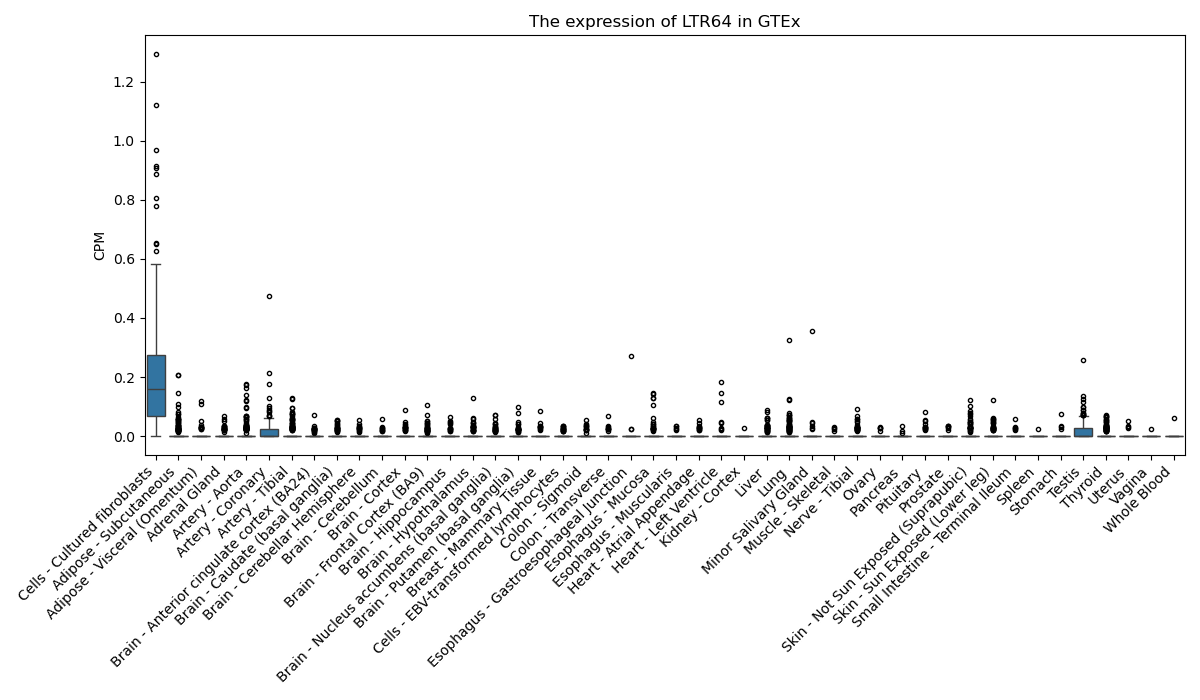
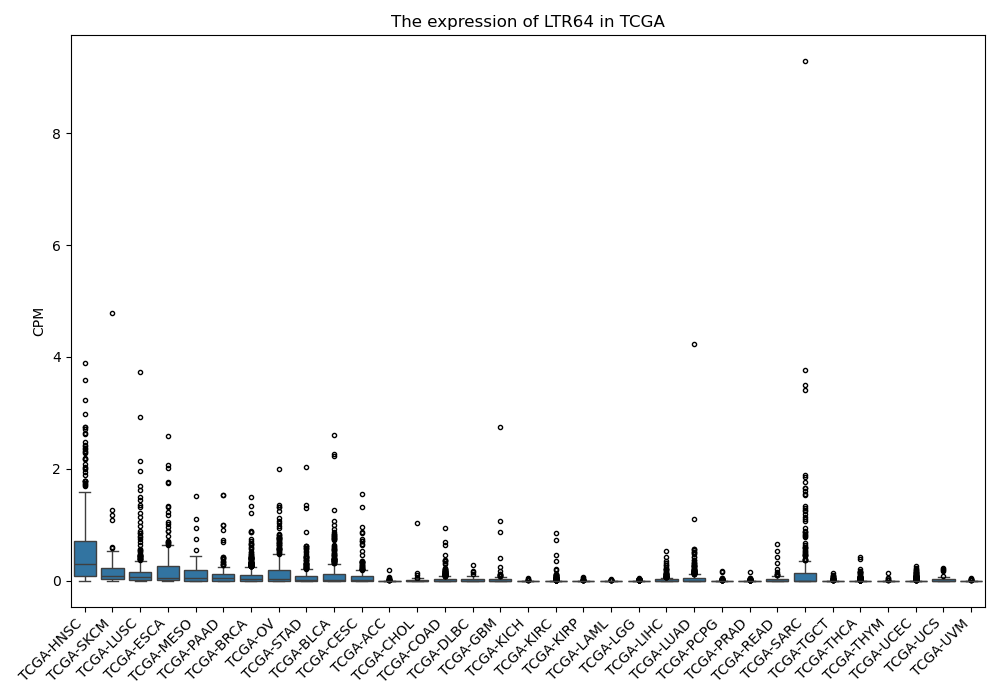
LTR64
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000563 |
|---|---|
| TE superfamily | ERV1 |
| TE class | LTR |
| Species | Primates |
| Length | 528 |
| Kimura value | 16.54 |
| Tau index | 1.0000 |
| Description | LTR64 (Long Terminal Repeat) for ERV1 endogenous retrovirus |
| Comment | 4 bp TSDs. |
| Sequence |
TGTGAGACAAAGTAGCAAACGTAAGAAGCCATGTTTGCTCATTTCTGCTTGCCAGCATAATTTCACAAAGCCCCTGACTCTGTGACGACGTGCAGCTCTCCGGAAAGATGCTTTGAAGACAAAACAGGATAGAGCACACGGCCCCCCACGTCTCTTGCCTGAGTCACTATATTCCTTAAAAGATAAATGACCCTAGTCCTTGCCTTTTCCTACACATAAGATAACGTCTGACGGGGTTAGTGATTATGCCTCTGTAATCTATAACCAGATGTACTCTTACACCCAAACCTTGATGTGATTCTGCTTTAATGTAACTTCTGAGCAAGTTTGATGTGATTTTGCACGTACTGAACCTCCACCACCTGTATATAAGCNGTGGGCTGAAACACTGCGCCGGAGCAGTCTGACAGAACCTCTCTGAAGGGCTGCTCCCGGGCTGTAGGCCTCGGTCTATAGTCCTCAGTAAGACTTCTGAATAAAACTAACTTTAATTCTTTAAAAGCTTGATTTTTTTTTCTTTAGTCGACA
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| LTR64 | GRF6 | 403 | 409 | + | 13.30 | TCTGACA |
| LTR64 | egrh-1 | 143 | 150 | + | 13.24 | CCCCCACG |
| LTR64 | PLAGL2 | 140 | 147 | + | 13.18 | GGCCCCCC |
| LTR64 | lin-32 | 359 | 366 | - | 13.17 | ACAGGTGG |
| LTR64 | IDD1 | 113 | 124 | - | 13.15 | TTTTGTCTTCAA |
| LTR64 | BATF3 | 160 | 166 | - | 13.14 | TGACTCA |
| LTR64 | dmrt99B | 4 | 16 | + | 13.13 | GAGACAAAGTAGC |
| LTR64 | JKD | 113 | 124 | - | 13.09 | TTTTGTCTTCAA |
| LTR64 | SNAI2 | 359 | 366 | + | 13.04 | CCACCTGT |
| LTR64 | DOF4.3 | 103 | 115 | + | 13.03 | GAAAGATGCTTTG |
TFBS enrichment in GRCh38
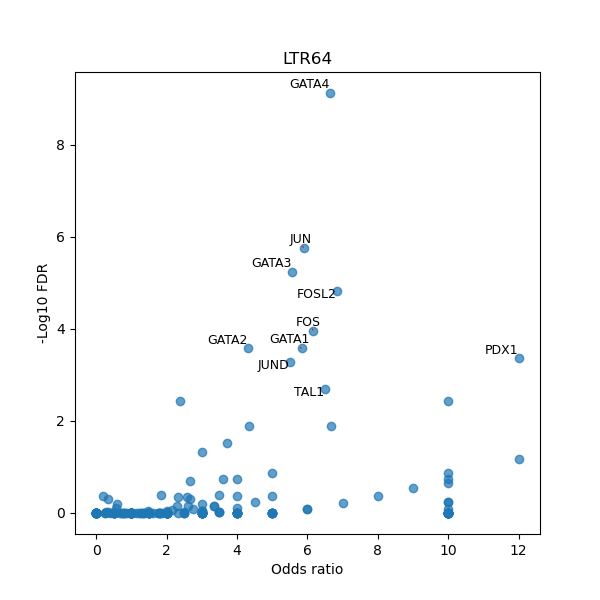
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.