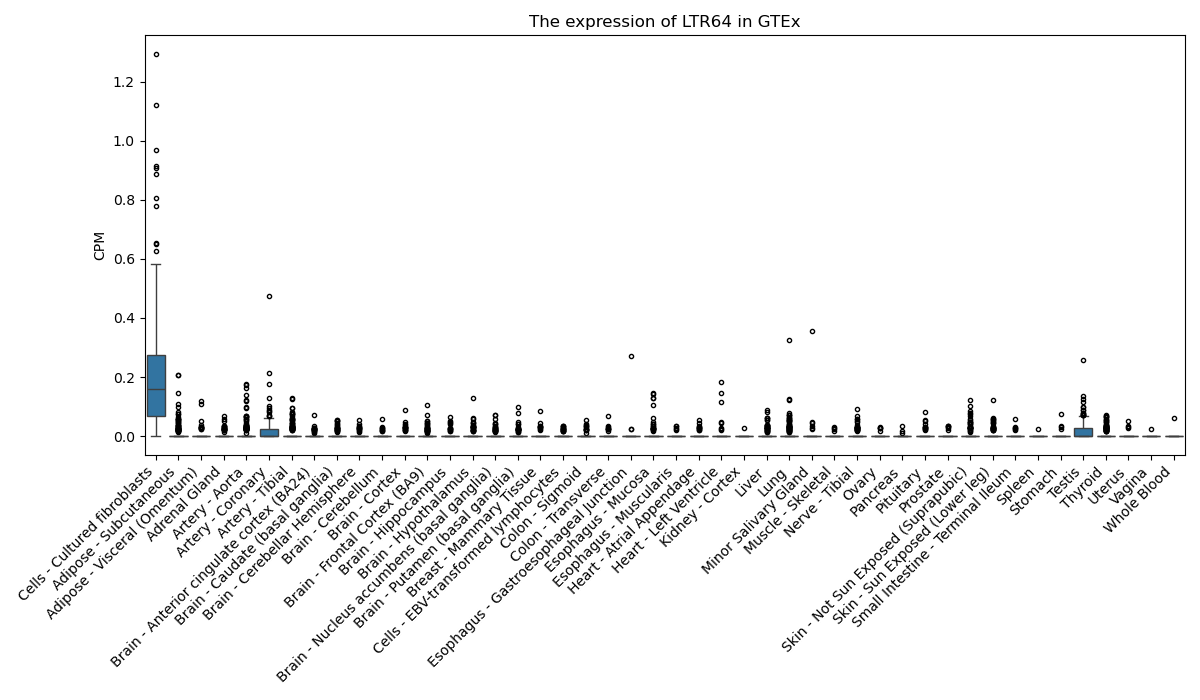
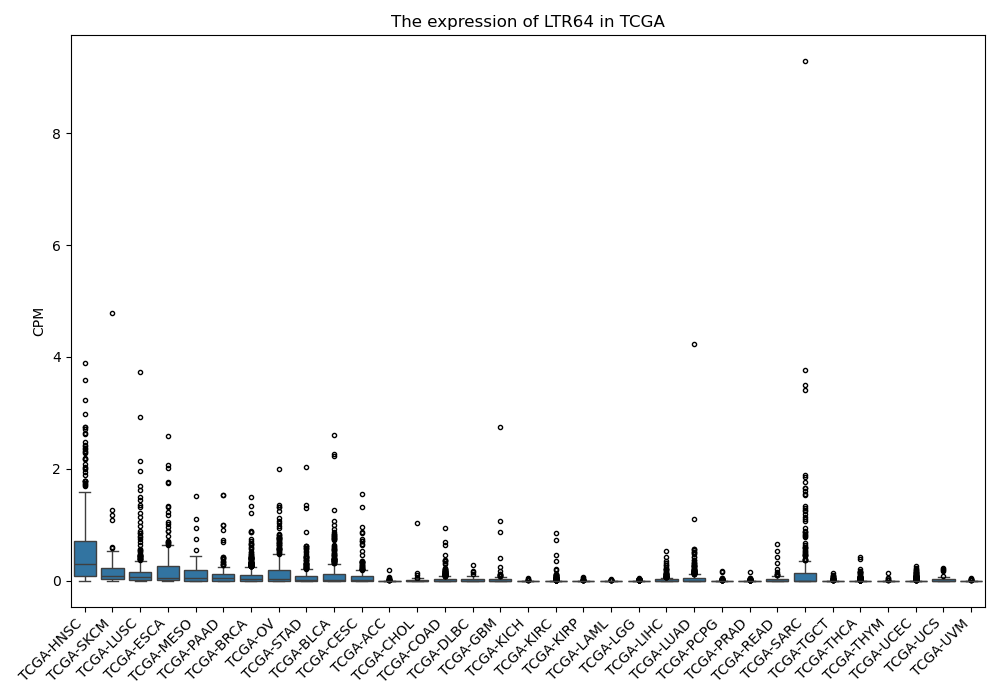
LTR64
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000563 |
|---|---|
| TE superfamily | ERV1 |
| TE class | LTR |
| Species | Primates |
| Length | 528 |
| Kimura value | 16.54 |
| Tau index | 1.0000 |
| Description | LTR64 (Long Terminal Repeat) for ERV1 endogenous retrovirus |
| Comment | 4 bp TSDs. |
| Sequence |
TGTGAGACAAAGTAGCAAACGTAAGAAGCCATGTTTGCTCATTTCTGCTTGCCAGCATAATTTCACAAAGCCCCTGACTCTGTGACGACGTGCAGCTCTCCGGAAAGATGCTTTGAAGACAAAACAGGATAGAGCACACGGCCCCCCACGTCTCTTGCCTGAGTCACTATATTCCTTAAAAGATAAATGACCCTAGTCCTTGCCTTTTCCTACACATAAGATAACGTCTGACGGGGTTAGTGATTATGCCTCTGTAATCTATAACCAGATGTACTCTTACACCCAAACCTTGATGTGATTCTGCTTTAATGTAACTTCTGAGCAAGTTTGATGTGATTTTGCACGTACTGAACCTCCACCACCTGTATATAAGCNGTGGGCTGAAACACTGCGCCGGAGCAGTCTGACAGAACCTCTCTGAAGGGCTGCTCCCGGGCTGTAGGCCTCGGTCTATAGTCCTCAGTAAGACTTCTGAATAAAACTAACTTTAATTCTTTAAAAGCTTGATTTTTTTTTCTTTAGTCGACA
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| LTR64 | Kah | 358 | 367 | + | 15.90 | ACCACCTGTA |
| LTR64 | RAP1 | 279 | 290 | - | 15.11 | AGGTTTGGGTGT |
| LTR64 | FOSL2 | 159 | 168 | - | 14.82 | AGTGACTCAG |
| LTR64 | ZNF768 | 412 | 420 | + | 14.62 | ACCTCTCTG |
| LTR64 | ASIL2 | 391 | 399 | - | 14.56 | CTCCGGCGC |
| LTR64 | TWIST1 | 265 | 272 | + | 14.54 | CCAGATGT |
| LTR64 | Mecom | 214 | 224 | + | 14.53 | ACATAAGATAA |
| LTR64 | DREB2F | 353 | 363 | + | 14.47 | CCTCCACCACC |
| LTR64 | MAC1 | 34 | 41 | + | 14.47 | TTTGCTCA |
| LTR64 | ENAP2 | 391 | 399 | - | 14.38 | CTCCGGCGC |
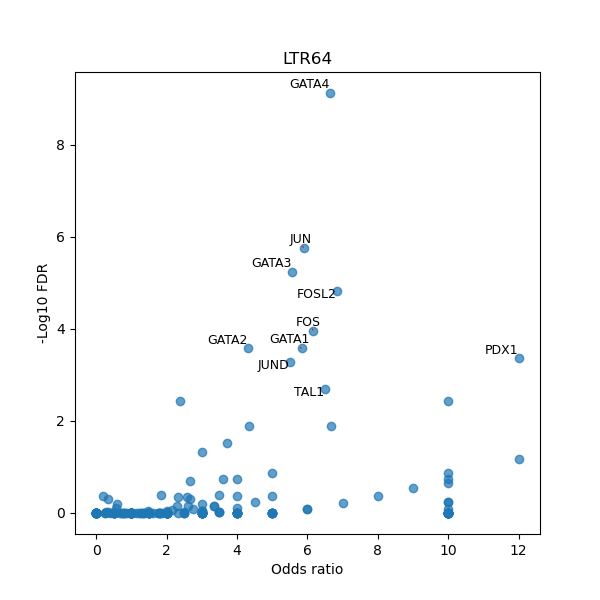
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.