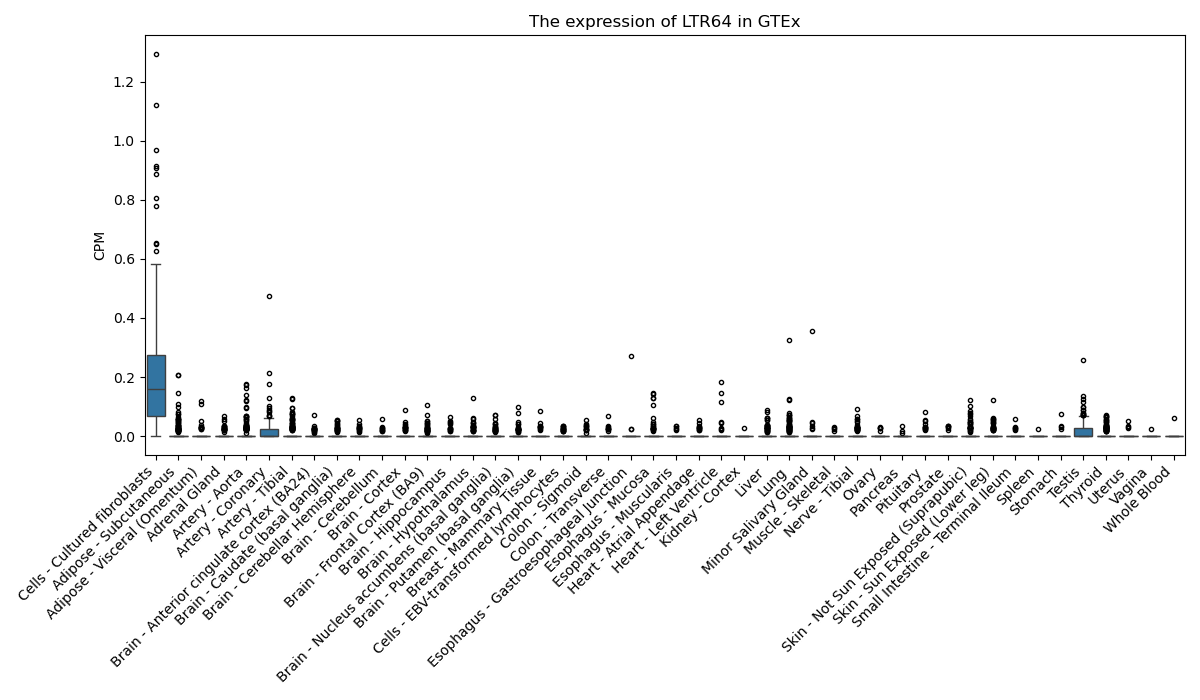
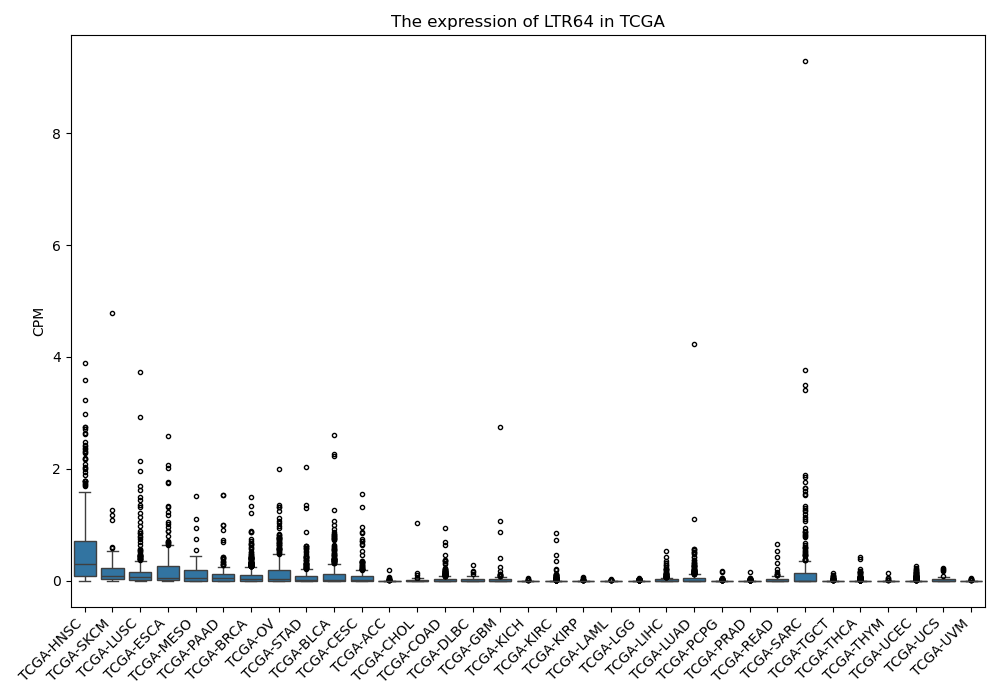
LTR64
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000563 |
|---|---|
| TE superfamily | ERV1 |
| TE class | LTR |
| Species | Primates |
| Length | 528 |
| Kimura value | 16.54 |
| Tau index | 1.0000 |
| Description | LTR64 (Long Terminal Repeat) for ERV1 endogenous retrovirus |
| Comment | 4 bp TSDs. |
| Sequence |
TGTGAGACAAAGTAGCAAACGTAAGAAGCCATGTTTGCTCATTTCTGCTTGCCAGCATAATTTCACAAAGCCCCTGACTCTGTGACGACGTGCAGCTCTCCGGAAAGATGCTTTGAAGACAAAACAGGATAGAGCACACGGCCCCCCACGTCTCTTGCCTGAGTCACTATATTCCTTAAAAGATAAATGACCCTAGTCCTTGCCTTTTCCTACACATAAGATAACGTCTGACGGGGTTAGTGATTATGCCTCTGTAATCTATAACCAGATGTACTCTTACACCCAAACCTTGATGTGATTCTGCTTTAATGTAACTTCTGAGCAAGTTTGATGTGATTTTGCACGTACTGAACCTCCACCACCTGTATATAAGCNGTGGGCTGAAACACTGCGCCGGAGCAGTCTGACAGAACCTCTCTGAAGGGCTGCTCCCGGGCTGTAGGCCTCGGTCTATAGTCCTCAGTAAGACTTCTGAATAAAACTAACTTTAATTCTTTAAAAGCTTGATTTTTTTTTCTTTAGTCGACA
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| LTR64 | erm | 34 | 44 | + | 14.35 | TTTGCTCATTT |
| LTR64 | sug | 140 | 151 | + | 14.28 | GGCCCCCCACGT |
| LTR64 | TB1 | 140 | 148 | + | 14.18 | GGCCCCCCA |
| LTR64 | DOF5.8 | 504 | 522 | + | 14.17 | TTGATTTTTTTTTCTTTAG |
| LTR64 | GLIS2 | 140 | 153 | + | 14.16 | GGCCCCCCACGTCT |
| LTR64 | snai-1 | 360 | 366 | + | 14.14 | CACCTGT |
| LTR64 | Zfx | 441 | 450 | - | 14.05 | ACCGAGGCCT |
| LTR64 | pha-4 | 32 | 41 | - | 13.98 | TGAGCAAACA |
| LTR64 | fos-1 | 160 | 166 | - | 13.76 | TGACTCA |
| LTR64 | Zm00001d027846 | 513 | 520 | - | 13.75 | AAAGAAAA |
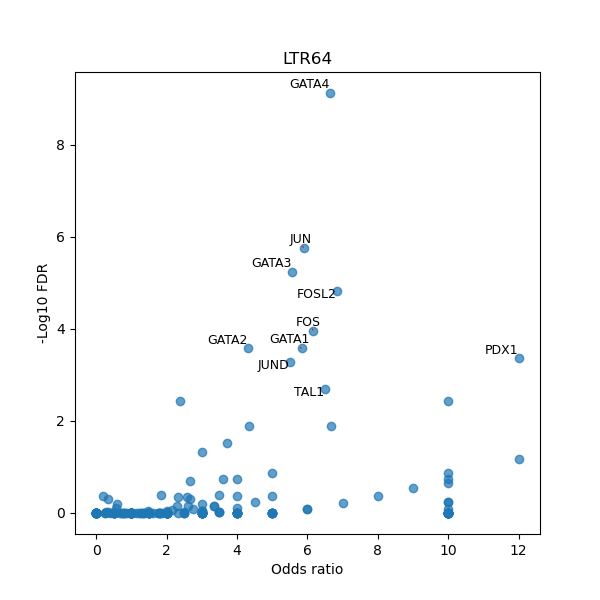
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.