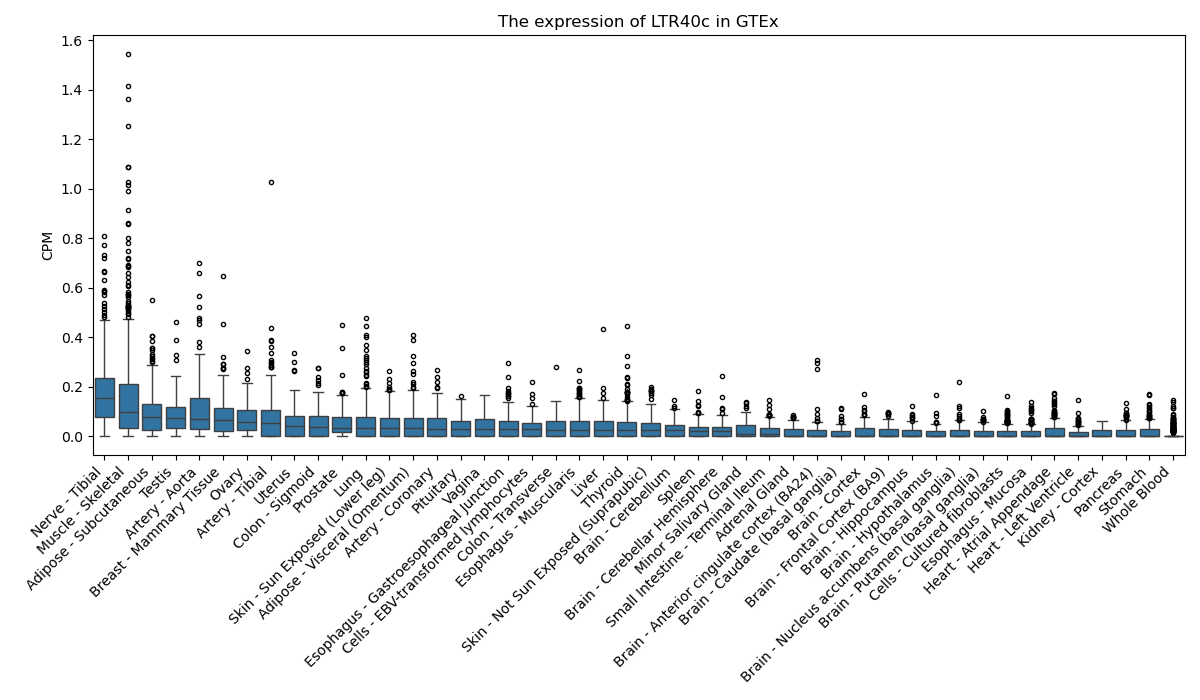
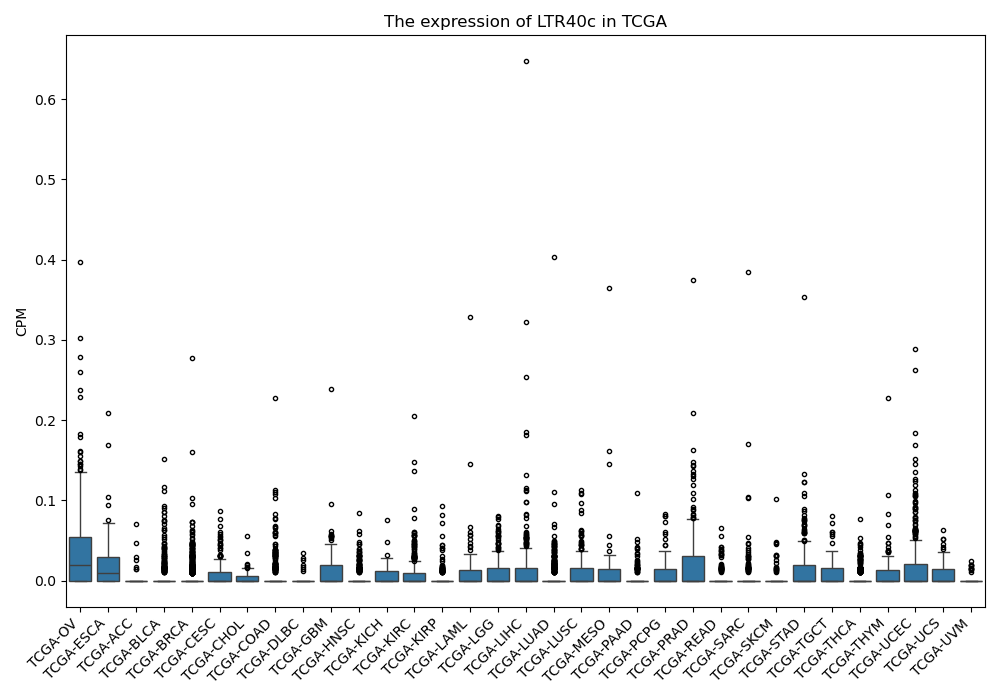
LTR40c
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000516 |
|---|---|
| TE superfamily | ERV3 |
| TE class | LTR |
| Species | Eutheria |
| Length | 465 |
| Kimura value | 24.54 |
| Tau index | 0.8479 |
| Description | LTR40b (Long Terminal Repeat) for HERVL40 endogenous retrovirus |
| Comment | 5 bp TSDs. |
| Sequence |
TGTTGGGAGACAATCCTCCATGGGCCCCTNGCGCTCCTGCACGTCTTGCTGGGTATGCCAAGAATGCAAGGCCCTGACCGCTCTTTACCTCGGGCCATTTCTCAGGGTTGTGTTTGCAGCGAGCAACCTTGAGGGATGAGGTAATGTCTCCCTCCGGGACAAAGAGCAGGCTTGCTTACTGCTTGCTATAAAAGCGGTGGATTCCCCAAGCTCAGTGTTCCTCNGCTGTAACGCAAACCCACTGCGTGCGCAGCATCCATCTGGGCCCTCCGCGTCGCCCCCGTGGGACTTGGGGGGNCANGGGGAACCGACGCAAACATGCTGATGCTCATGCTGCTTGCTGTGCCGTGAGTAATAAAGTCCTTTGTCTCTGACCCAGGAGTCTCGTGTCTTCTGCCAGCATCCATGAAACAGTAACAGGCTAACTTATTAGCTTGTAAGTAGGGTAAAATCCCAGACCTGACA
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| LTR40c | NEUROG2 | 257 | 263 | - | 13.29 | CAGATGG |
| LTR40c | Neurod2 | 257 | 264 | - | 13.17 | CCAGATGG |
| LTR40c | RELB | 201 | 207 | + | 13.14 | ATTCCCC |
| LTR40c | ATF2 | 136 | 145 | + | 13.06 | ATGAGGTAAT |
| LTR40c | Mad | 267 | 281 | - | 13.00 | GGGGCGACGCGGAGG |
| LTR40c | AT5G47660 | 445 | 451 | + | 13.00 | GGTAAAA |
| LTR40c | LBD13 | 194 | 201 | - | 12.94 | TCCACCGC |
| LTR40c | ARF25 | 144 | 155 | - | 12.90 | GGAGGGAGACAT |
| LTR40c | SATB1 | 427 | 433 | - | 12.89 | CTAATAA |
| LTR40c | RIM101 | 56 | 62 | + | 12.82 | TGCCAAG |
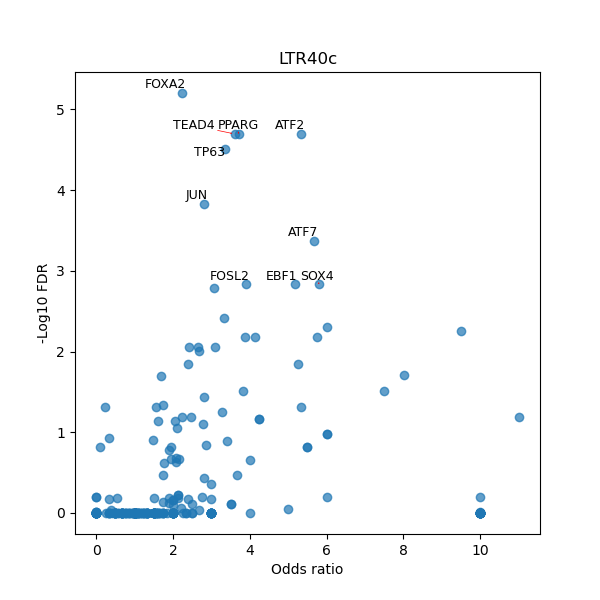
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.