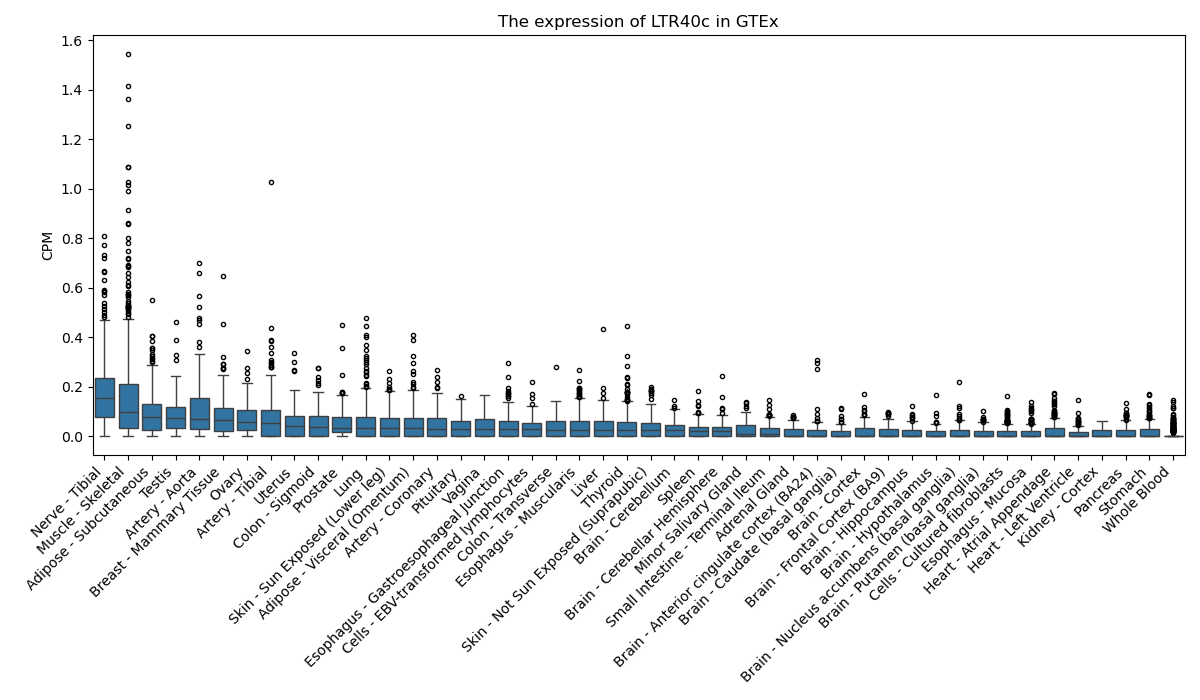
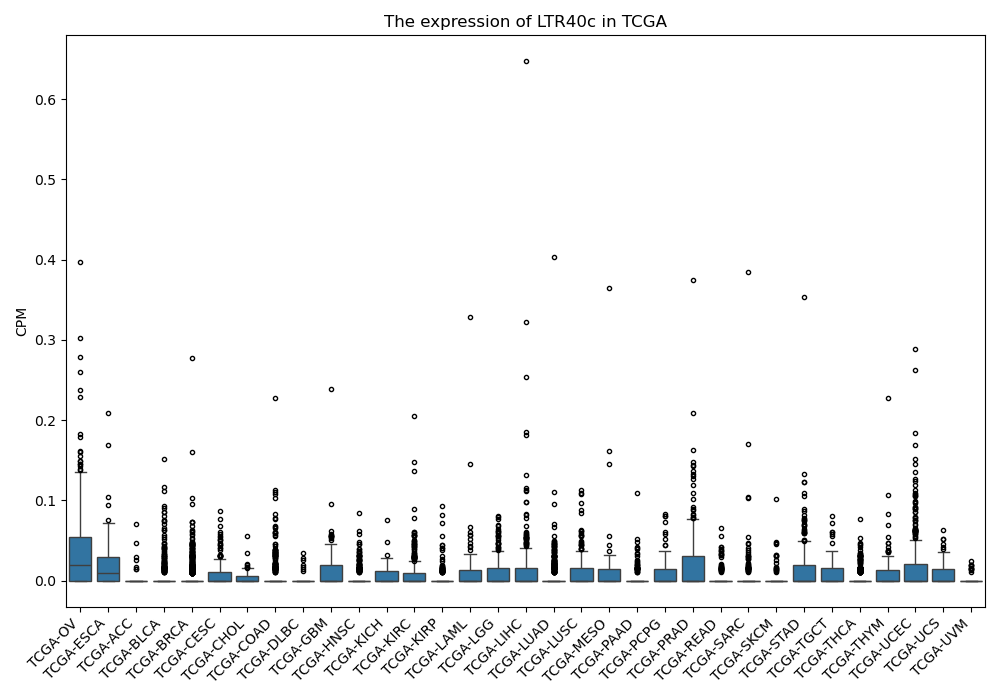
LTR40c
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000516 |
|---|---|
| TE superfamily | ERV3 |
| TE class | LTR |
| Species | Eutheria |
| Length | 465 |
| Kimura value | 24.54 |
| Tau index | 0.8479 |
| Description | LTR40b (Long Terminal Repeat) for HERVL40 endogenous retrovirus |
| Comment | 5 bp TSDs. |
| Sequence |
TGTTGGGAGACAATCCTCCATGGGCCCCTNGCGCTCCTGCACGTCTTGCTGGGTATGCCAAGAATGCAAGGCCCTGACCGCTCTTTACCTCGGGCCATTTCTCAGGGTTGTGTTTGCAGCGAGCAACCTTGAGGGATGAGGTAATGTCTCCCTCCGGGACAAAGAGCAGGCTTGCTTACTGCTTGCTATAAAAGCGGTGGATTCCCCAAGCTCAGTGTTCCTCNGCTGTAACGCAAACCCACTGCGTGCGCAGCATCCATCTGGGCCCTCCGCGTCGCCCCCGTGGGACTTGGGGGGNCANGGGGAACCGACGCAAACATGCTGATGCTCATGCTGCTTGCTGTGCCGTGAGTAATAAAGTCCTTTGTCTCTGACCCAGGAGTCTCGTGTCTTCTGCCAGCATCCATGAAACAGTAACAGGCTAACTTATTAGCTTGTAAGTAGGGTAAAATCCCAGACCTGACA
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| LTR40c | JUND | 135 | 145 | + | 15.30 | GATGAGGTAAT |
| LTR40c | NFkb | 197 | 207 | - | 15.25 | GGGGAATCCAC |
| LTR40c | Sox14 | 360 | 370 | + | 14.85 | GTCCTTTGTCT |
| LTR40c | Sox14 | 157 | 167 | - | 14.72 | GCTCTTTGTCC |
| LTR40c | ZNF692 | 261 | 268 | - | 14.58 | GGGCCCAG |
| LTR40c | JUN | 136 | 145 | + | 14.53 | ATGAGGTAAT |
| LTR40c | ARF10 | 145 | 155 | - | 14.28 | GGAGGGAGACA |
| LTR40c | Mad | 264 | 278 | - | 14.28 | GCGACGCGGAGGGCC |
| LTR40c | egrh-1 | 276 | 283 | + | 14.01 | CGCCCCCG |
| LTR40c | PK08602.1 | 193 | 200 | - | 13.89 | CCACCGCT |
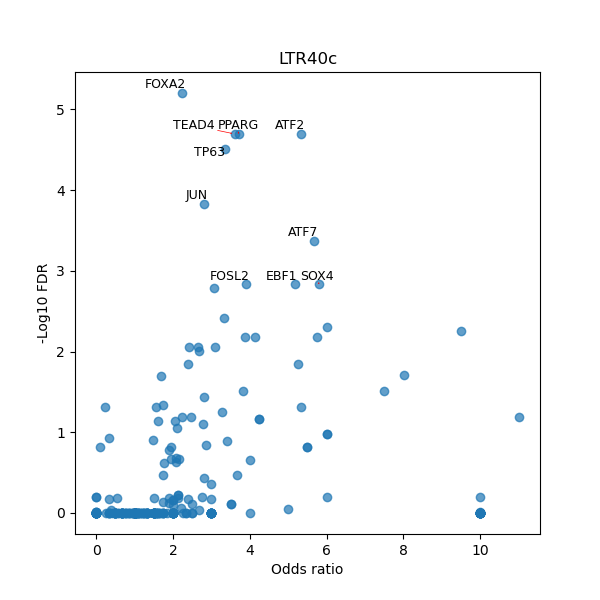
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.