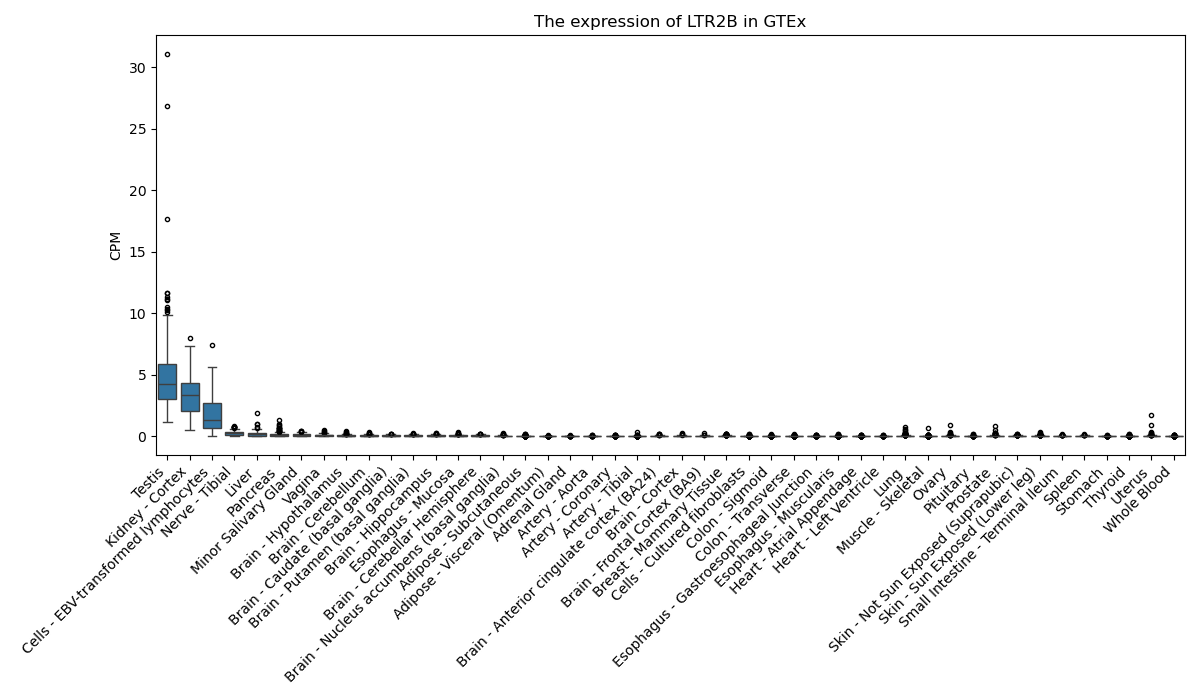
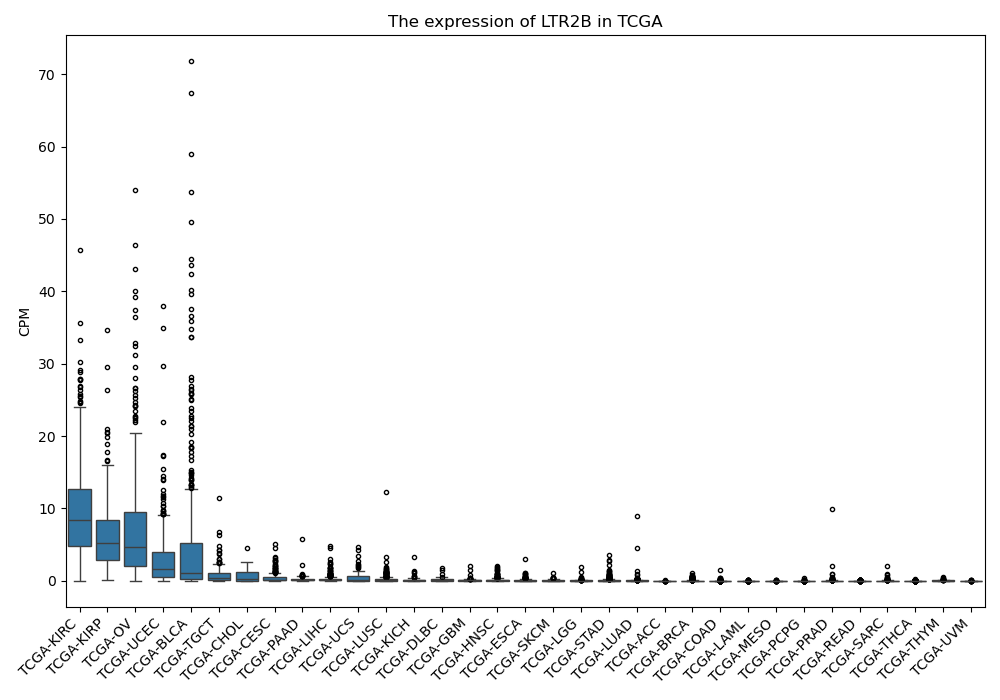
LTR2B
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000484 |
|---|---|
| TE superfamily | ERV1 |
| TE class | LTR |
| Species | Hominoidea |
| Length | 483 |
| Kimura value | 5.21 |
| Tau index | 0.9714 |
| Description | LTR2B (Long Terminal Repeat) for Harlequin endogenous retrovirus |
| Comment | - |
| Sequence |
TGAGGGAAGAGAGAGACCCTCTCATATTGTTTTATATTGTTTTATACTCAGTACCTGTTTTAAGAAAAAAACAACAAGGAAGTAAAACCAAAGACAGGCAGCCCGGCGCCAGGCCCGAAACCAGGCCTGGGCCTGCCTGGCCTAAACCCAGTAGTTAAAAATCAACTCATGACTTAGAAACCGATGTTATTCATAGATTCCAGACATTGTATAGAAGAACATTGTGAAACTCCCTGCCCTGTTCTGTTTCTCTCTGACCACCGGTGCATGCAGCCCCTGTCACGTACCCCCTGCTTGCTCAAATCAATCACGACCCTTTCATGTGAAATCTTTAGTGTTGTGAGCCCTTAAAAGGGACAGAAATTGTGCACTCGGGGAGCTCGGATTTTAAGGCAGTAGCTTGCCGATGCTCCCAGCTGAATAAAGCCCTTCCTTCTACAACTCGGTGTCTGAGAGGTTTTGTCTGCGGCTCGTCCTGCTACA
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| LTR2B | IRF5 | 115 | 128 | + | 12.43 | CCGAAACCAGGCCT |
| LTR2B | STP1 | 465 | 472 | + | 12.41 | TGCGGCTC |
| LTR2B | ERF::FOXO1 | 90 | 101 | + | 12.41 | AAAGACAGGCAG |
| LTR2B | IDD4 | 63 | 77 | + | 12.35 | AGAAAAAAACAACAA |
| LTR2B | CEBPA | 221 | 230 | - | 12.30 | GTTTCACAAT |
| LTR2B | Trl | 9 | 17 | + | 12.19 | GAGAGAGAC |
| LTR2B | RAMOSA1 | 3 | 16 | + | 12.14 | AGGGAAGAGAGAGA |
| LTR2B | fkh-9 | 67 | 74 | + | 12.13 | AAAAACAA |
| LTR2B | CTCF | 284 | 298 | - | 12.07 | GCAAGCAGGGGGTAC |
| LTR2B | ERF109 | 103 | 110 | + | 12.05 | CCGGCGCC |
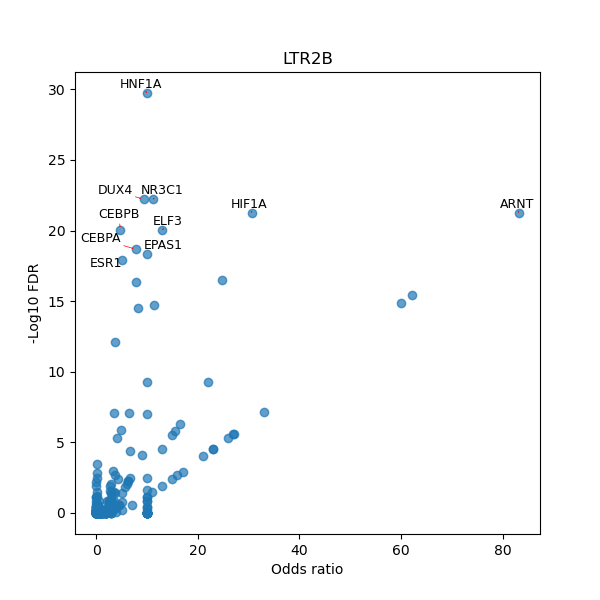
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.