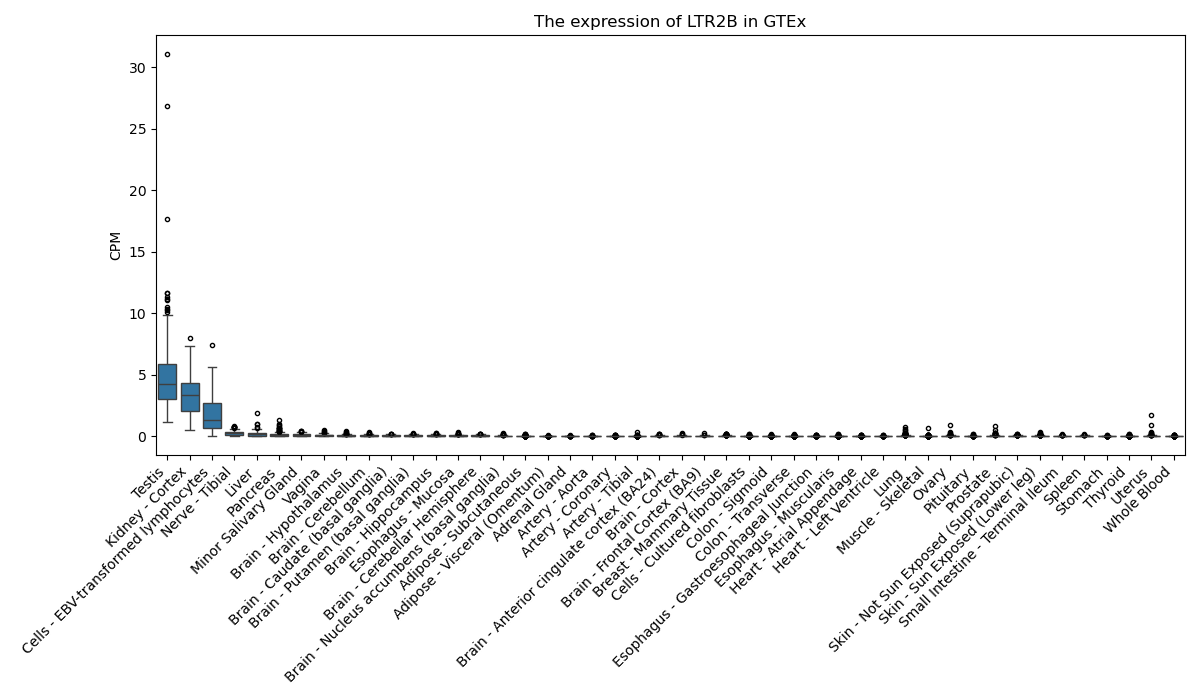
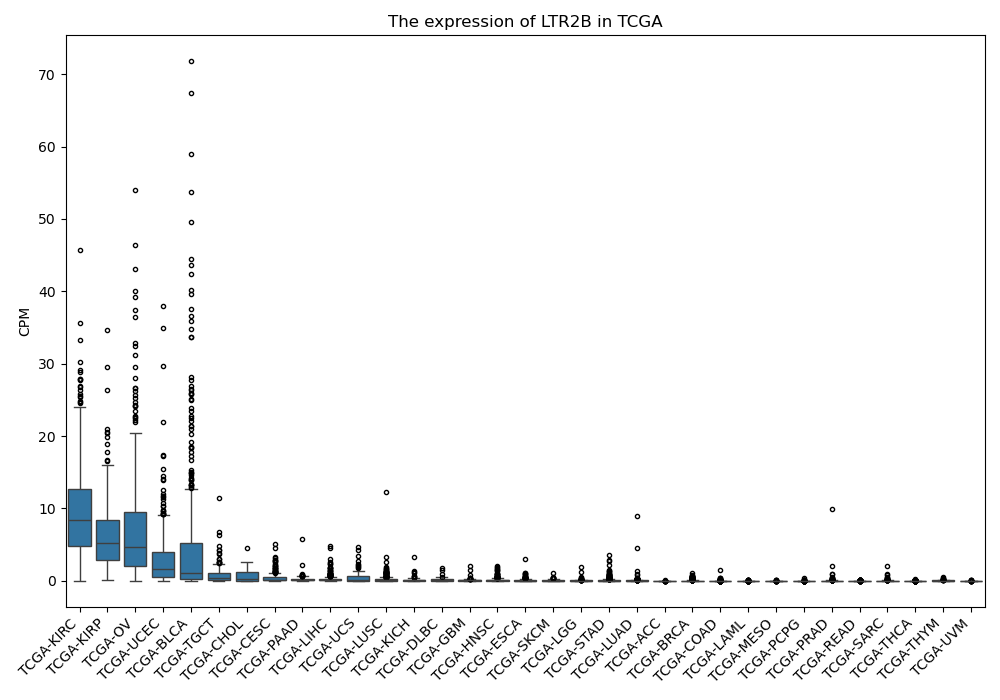
LTR2B
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000484 |
|---|---|
| TE superfamily | ERV1 |
| TE class | LTR |
| Species | Hominoidea |
| Length | 483 |
| Kimura value | 5.21 |
| Tau index | 0.9714 |
| Description | LTR2B (Long Terminal Repeat) for Harlequin endogenous retrovirus |
| Comment | - |
| Sequence |
TGAGGGAAGAGAGAGACCCTCTCATATTGTTTTATATTGTTTTATACTCAGTACCTGTTTTAAGAAAAAAACAACAAGGAAGTAAAACCAAAGACAGGCAGCCCGGCGCCAGGCCCGAAACCAGGCCTGGGCCTGCCTGGCCTAAACCCAGTAGTTAAAAATCAACTCATGACTTAGAAACCGATGTTATTCATAGATTCCAGACATTGTATAGAAGAACATTGTGAAACTCCCTGCCCTGTTCTGTTTCTCTCTGACCACCGGTGCATGCAGCCCCTGTCACGTACCCCCTGCTTGCTCAAATCAATCACGACCCTTTCATGTGAAATCTTTAGTGTTGTGAGCCCTTAAAAGGGACAGAAATTGTGCACTCGGGGAGCTCGGATTTTAAGGCAGTAGCTTGCCGATGCTCCCAGCTGAATAAAGCCCTTCCTTCTACAACTCGGTGTCTGAGAGGTTTTGTCTGCGGCTCGTCCTGCTACA
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| LTR2B | Zfx | 123 | 132 | - | 15.80 | GCCCAGGCCT |
| LTR2B | ZNF667 | 341 | 351 | - | 15.80 | TTAAGGGCTCA |
| LTR2B | Trl | 7 | 15 | + | 15.63 | AAGAGAGAG |
| LTR2B | Zfx | 125 | 134 | + | 15.36 | GCCTGGGCCT |
| LTR2B | ZNF417 | 105 | 111 | + | 14.55 | GGCGCCA |
| LTR2B | ZNF213 | 121 | 132 | - | 14.31 | GCCCAGGCCTGG |
| LTR2B | Elf5 | 76 | 83 | + | 14.24 | AAGGAAGT |
| LTR2B | Os05g0497200 | 104 | 111 | + | 14.24 | CGGCGCCA |
| LTR2B | GLYMA-08G357600 | 264 | 272 | + | 14.08 | GTGCATGCA |
| LTR2B | brk | 105 | 112 | - | 13.61 | CTGGCGCC |
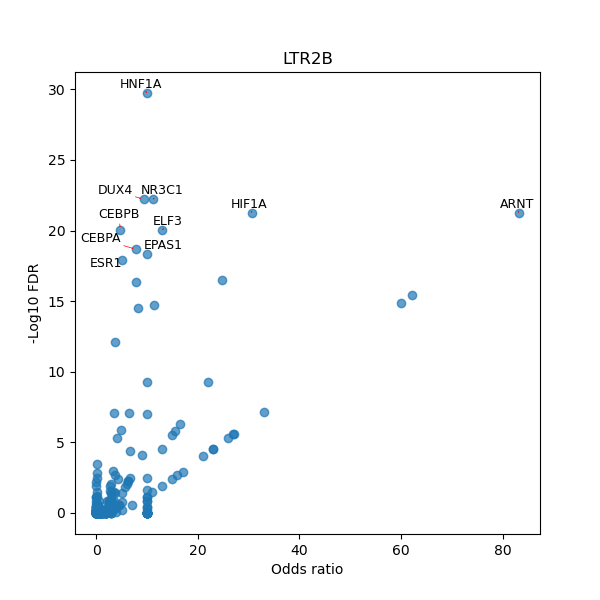
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.