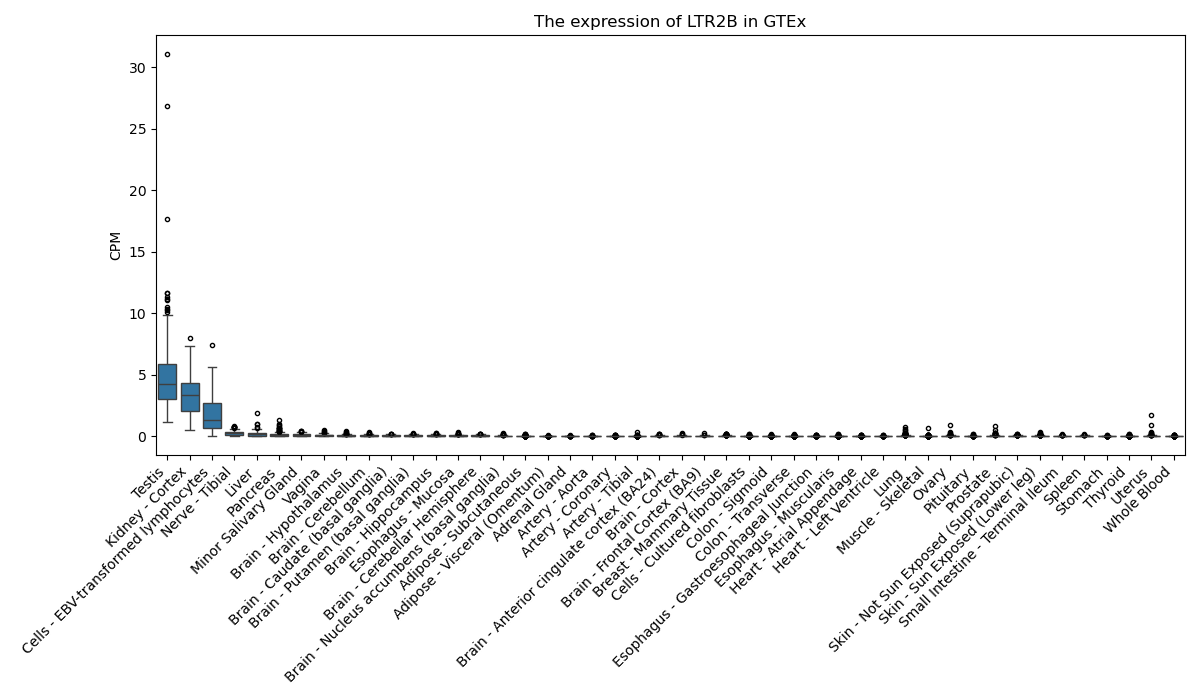
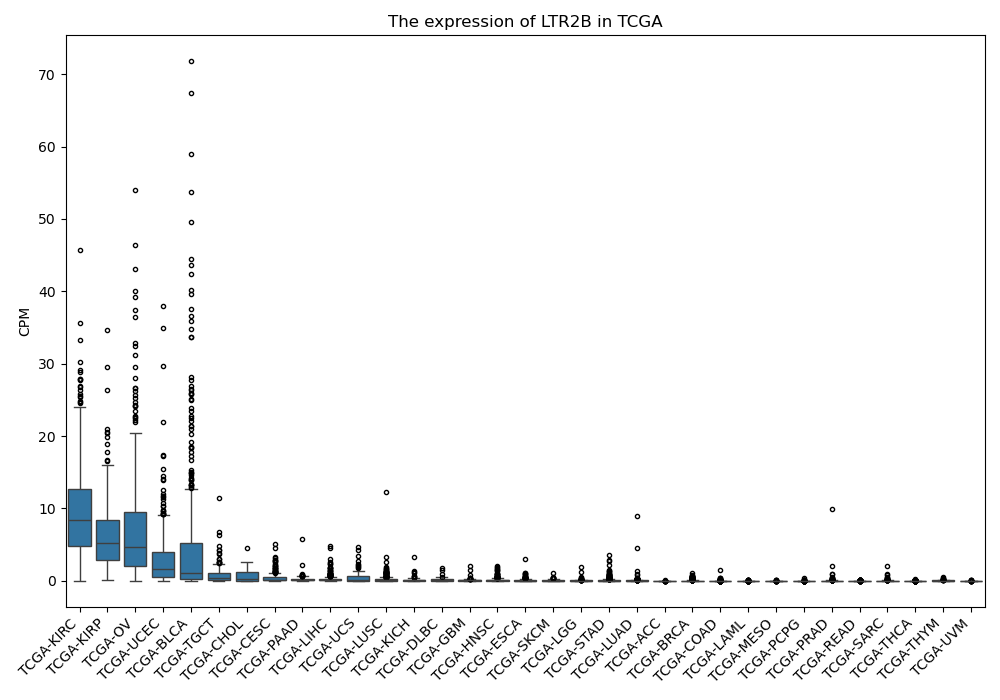
LTR2B
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000484 |
|---|---|
| TE superfamily | ERV1 |
| TE class | LTR |
| Species | Hominoidea |
| Length | 483 |
| Kimura value | 5.21 |
| Tau index | 0.9714 |
| Description | LTR2B (Long Terminal Repeat) for Harlequin endogenous retrovirus |
| Comment | - |
| Sequence |
TGAGGGAAGAGAGAGACCCTCTCATATTGTTTTATATTGTTTTATACTCAGTACCTGTTTTAAGAAAAAAACAACAAGGAAGTAAAACCAAAGACAGGCAGCCCGGCGCCAGGCCCGAAACCAGGCCTGGGCCTGCCTGGCCTAAACCCAGTAGTTAAAAATCAACTCATGACTTAGAAACCGATGTTATTCATAGATTCCAGACATTGTATAGAAGAACATTGTGAAACTCCCTGCCCTGTTCTGTTTCTCTCTGACCACCGGTGCATGCAGCCCCTGTCACGTACCCCCTGCTTGCTCAAATCAATCACGACCCTTTCATGTGAAATCTTTAGTGTTGTGAGCCCTTAAAAGGGACAGAAATTGTGCACTCGGGGAGCTCGGATTTTAAGGCAGTAGCTTGCCGATGCTCCCAGCTGAATAAAGCCCTTCCTTCTACAACTCGGTGTCTGAGAGGTTTTGTCTGCGGCTCGTCCTGCTACA
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| LTR2B | BCL11A | 254 | 260 | + | 12.85 | CTGACCA |
| LTR2B | MEIS1 | 302 | 310 | + | 12.76 | AATCAATCA |
| LTR2B | TCF7L2 | 316 | 324 | - | 12.75 | ACATGAAAG |
| LTR2B | SoxB1 | 215 | 225 | + | 12.56 | AAGAACATTGT |
| LTR2B | BAD1 | 102 | 113 | + | 12.51 | CCCGGCGCCAGG |
| LTR2B | HRS1 | 190 | 201 | + | 12.50 | TTCATAGATTCC |
| LTR2B | Hnf1A | 315 | 324 | + | 12.48 | CCTTTCATGT |
| LTR2B | bs | 346 | 355 | + | 12.44 | CCTTAAAAGG |
| LTR2B | dsx | 204 | 210 | - | 12.44 | ACAATGT |
| LTR2B | dsx | 219 | 225 | - | 12.44 | ACAATGT |
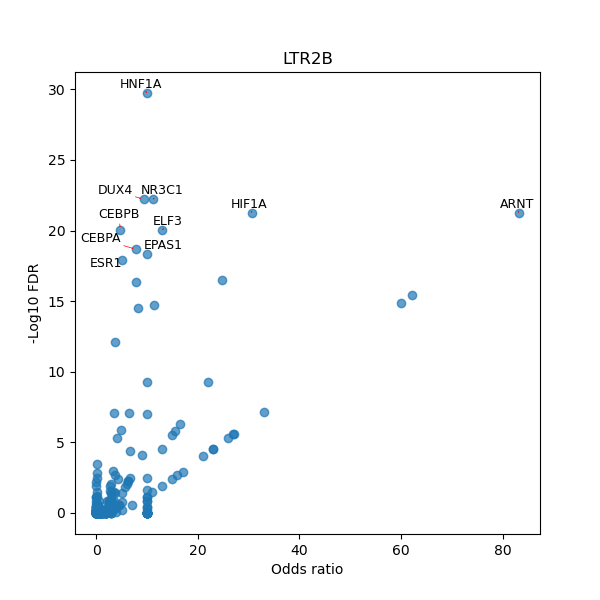
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.