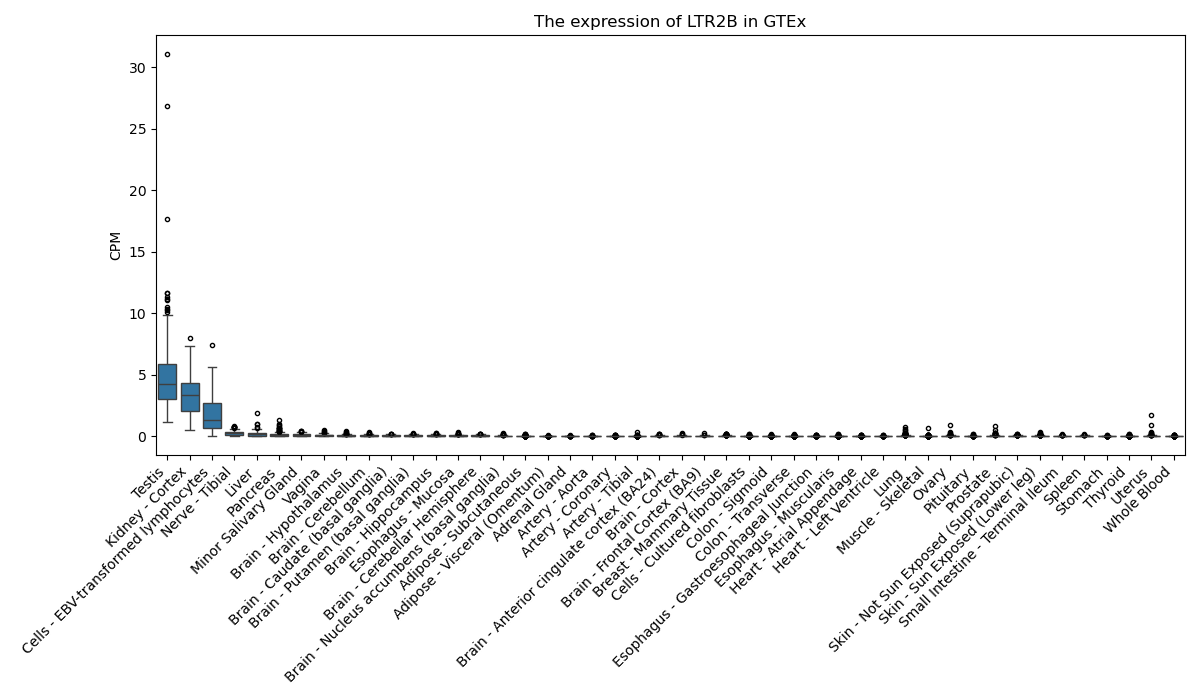
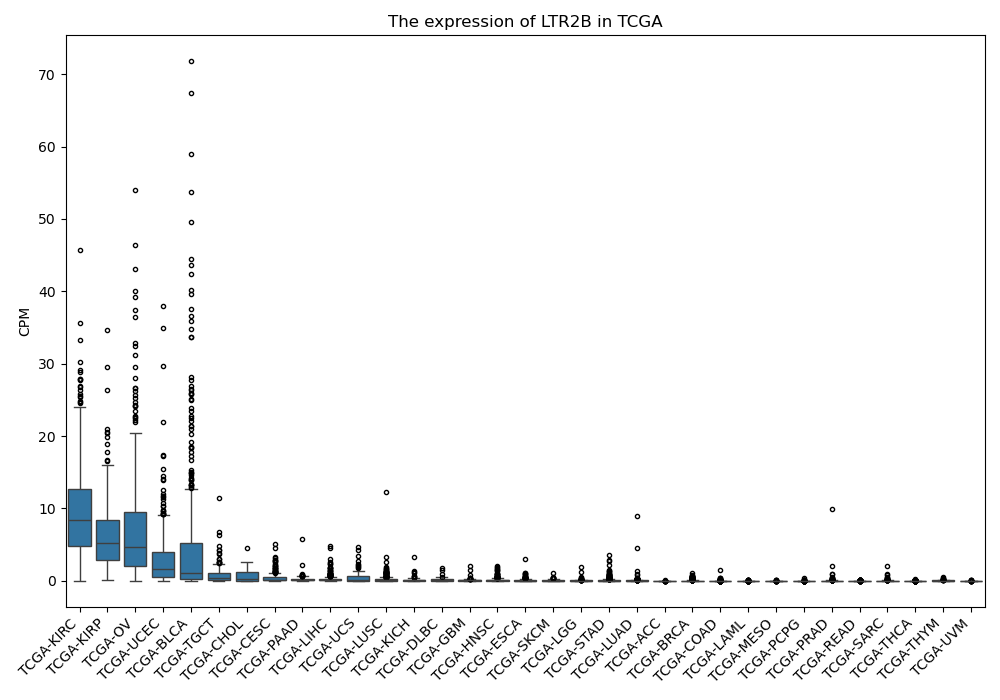
LTR2B
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000484 |
|---|---|
| TE superfamily | ERV1 |
| TE class | LTR |
| Species | Hominoidea |
| Length | 483 |
| Kimura value | 5.21 |
| Tau index | 0.9714 |
| Description | LTR2B (Long Terminal Repeat) for Harlequin endogenous retrovirus |
| Comment | - |
| Sequence |
TGAGGGAAGAGAGAGACCCTCTCATATTGTTTTATATTGTTTTATACTCAGTACCTGTTTTAAGAAAAAAACAACAAGGAAGTAAAACCAAAGACAGGCAGCCCGGCGCCAGGCCCGAAACCAGGCCTGGGCCTGCCTGGCCTAAACCCAGTAGTTAAAAATCAACTCATGACTTAGAAACCGATGTTATTCATAGATTCCAGACATTGTATAGAAGAACATTGTGAAACTCCCTGCCCTGTTCTGTTTCTCTCTGACCACCGGTGCATGCAGCCCCTGTCACGTACCCCCTGCTTGCTCAAATCAATCACGACCCTTTCATGTGAAATCTTTAGTGTTGTGAGCCCTTAAAAGGGACAGAAATTGTGCACTCGGGGAGCTCGGATTTTAAGGCAGTAGCTTGCCGATGCTCCCAGCTGAATAAAGCCCTTCCTTCTACAACTCGGTGTCTGAGAGGTTTTGTCTGCGGCTCGTCCTGCTACA
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| LTR2B | ZNF213 | 131 | 142 | - | 13.50 | GGCCAGGCAGGC |
| LTR2B | brk | 103 | 110 | + | 13.31 | CCGGCGCC |
| LTR2B | ZBED2 | 116 | 122 | + | 13.20 | CGAAACC |
| LTR2B | SFL1 | 211 | 220 | + | 13.20 | ATAGAAGAAC |
| LTR2B | sma-4 | 200 | 206 | - | 13.18 | TGTCTGG |
| LTR2B | Hmga1 | 155 | 162 | - | 12.99 | ATTTTTAA |
| LTR2B | Hand1::Tcf3 | 197 | 205 | - | 12.99 | GTCTGGAAT |
| LTR2B | GLYMA-08G357600 | 265 | 273 | - | 12.96 | CTGCATGCA |
| LTR2B | HMRA1 | 363 | 369 | - | 12.91 | GCACAAT |
| LTR2B | Stat2 | 241 | 250 | - | 12.87 | GAAACAGAAC |
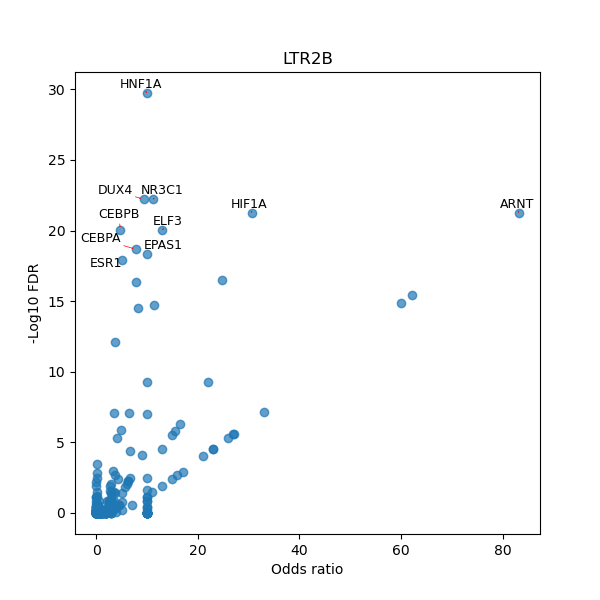
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.