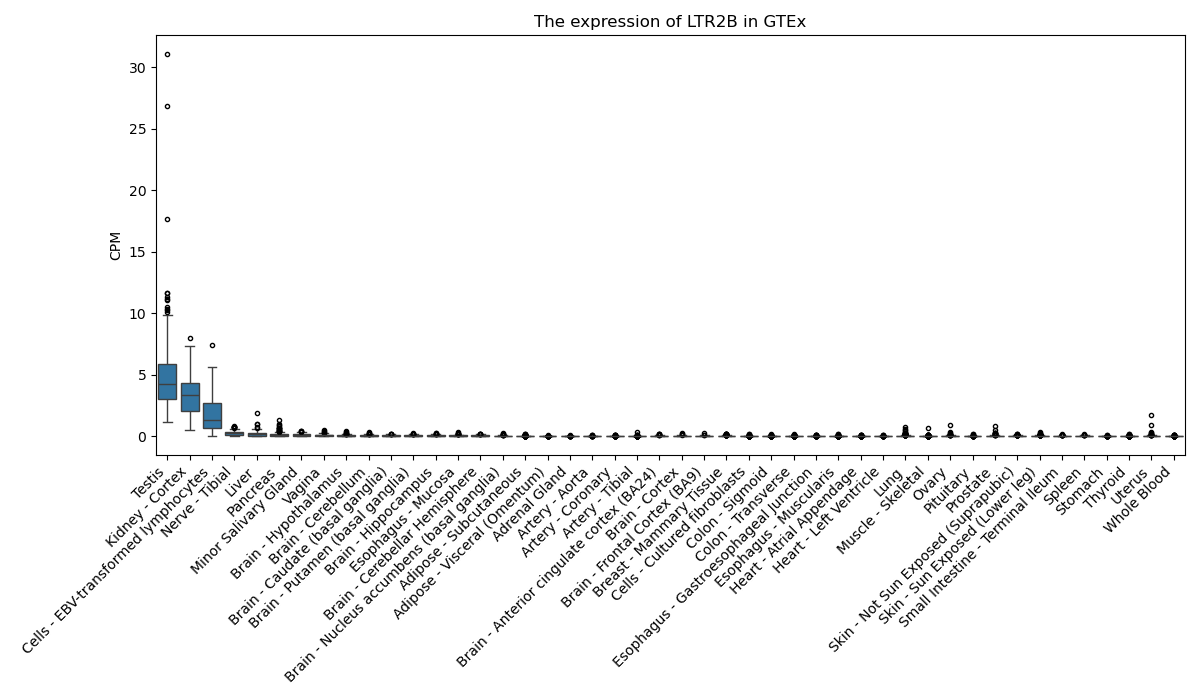
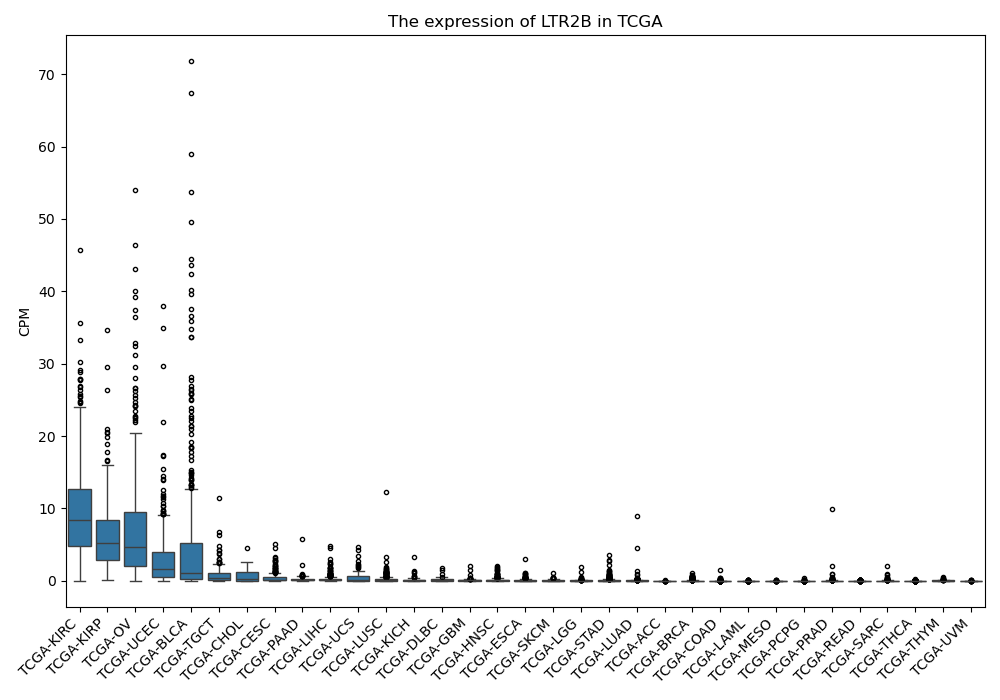
LTR2B
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000484 |
|---|---|
| TE superfamily | ERV1 |
| TE class | LTR |
| Species | Hominoidea |
| Length | 483 |
| Kimura value | 5.21 |
| Tau index | 0.9714 |
| Description | LTR2B (Long Terminal Repeat) for Harlequin endogenous retrovirus |
| Comment | - |
| Sequence |
TGAGGGAAGAGAGAGACCCTCTCATATTGTTTTATATTGTTTTATACTCAGTACCTGTTTTAAGAAAAAAACAACAAGGAAGTAAAACCAAAGACAGGCAGCCCGGCGCCAGGCCCGAAACCAGGCCTGGGCCTGCCTGGCCTAAACCCAGTAGTTAAAAATCAACTCATGACTTAGAAACCGATGTTATTCATAGATTCCAGACATTGTATAGAAGAACATTGTGAAACTCCCTGCCCTGTTCTGTTTCTCTCTGACCACCGGTGCATGCAGCCCCTGTCACGTACCCCCTGCTTGCTCAAATCAATCACGACCCTTTCATGTGAAATCTTTAGTGTTGTGAGCCCTTAAAAGGGACAGAAATTGTGCACTCGGGGAGCTCGGATTTTAAGGCAGTAGCTTGCCGATGCTCCCAGCTGAATAAAGCCCTTCCTTCTACAACTCGGTGTCTGAGAGGTTTTGTCTGCGGCTCGTCCTGCTACA
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| LTR2B | ZNF16 | 398 | 418 | - | 6.73 | AGCTGGGAGCATCGGCAAGCT |
| LTR2B | TP53 | 120 | 137 | + | 6.55 | ACCAGGCCTGGGCCTGCC |
| LTR2B | TP53 | 120 | 137 | - | 6.18 | GGCAGGCCCAGGCCTGGT |
| LTR2B | RAX3 | 280 | 291 | - | 6.02 | GGGGGTACGTGA |
| LTR2B | RAMOSA1 | 247 | 260 | - | 5.84 | TGGTCAGAGAGAAA |
| LTR2B | BPC1 | 229 | 252 | - | 4.62 | GAGAAACAGAACAGGGCAGGGAGT |
| LTR2B | Poxm | 259 | 273 | + | 0.02 | CACCGGTGCATGCAG |
| LTR2B | BPC5 | 224 | 253 | - | -36.59 | AGAGAAACAGAACAGGGCAGGGAGTTTCAC |
| LTR2B | BPC5 | 226 | 255 | - | -46.88 | AGAGAGAAACAGAACAGGGCAGGGAGTTTC |
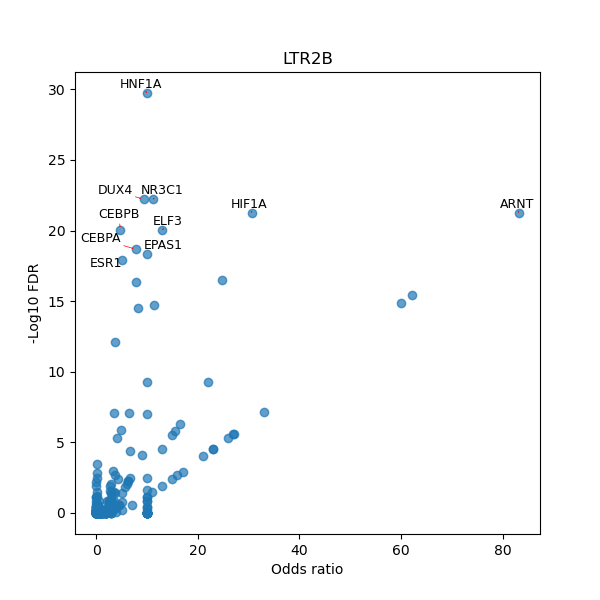
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.