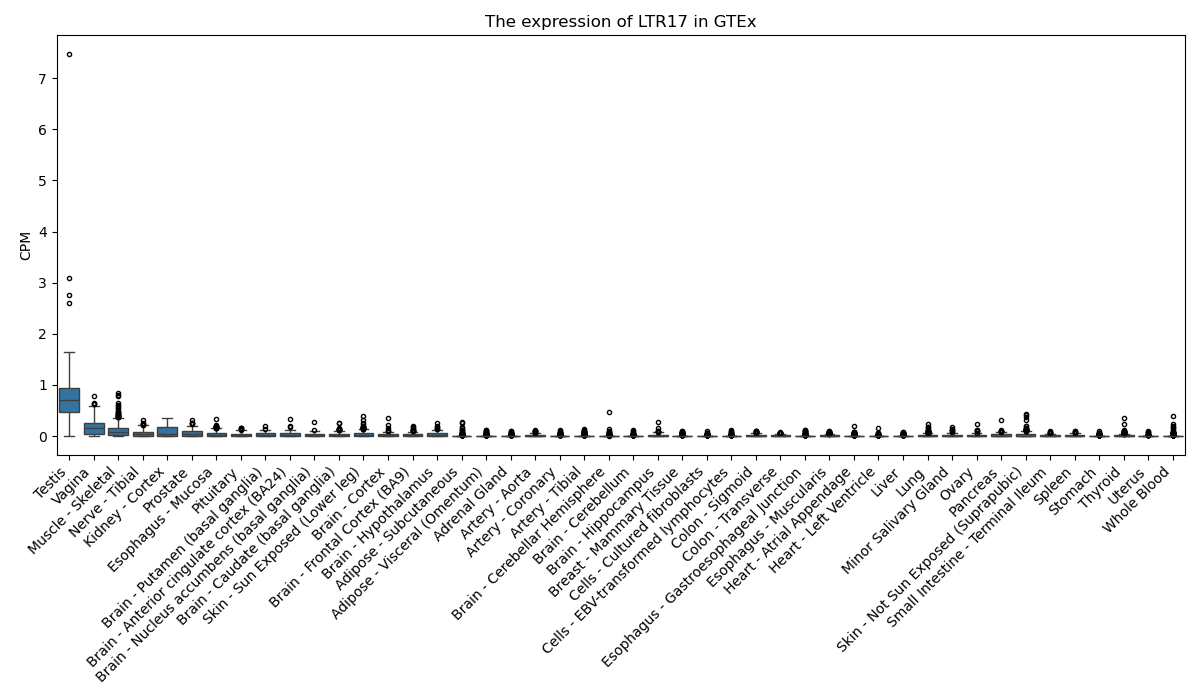
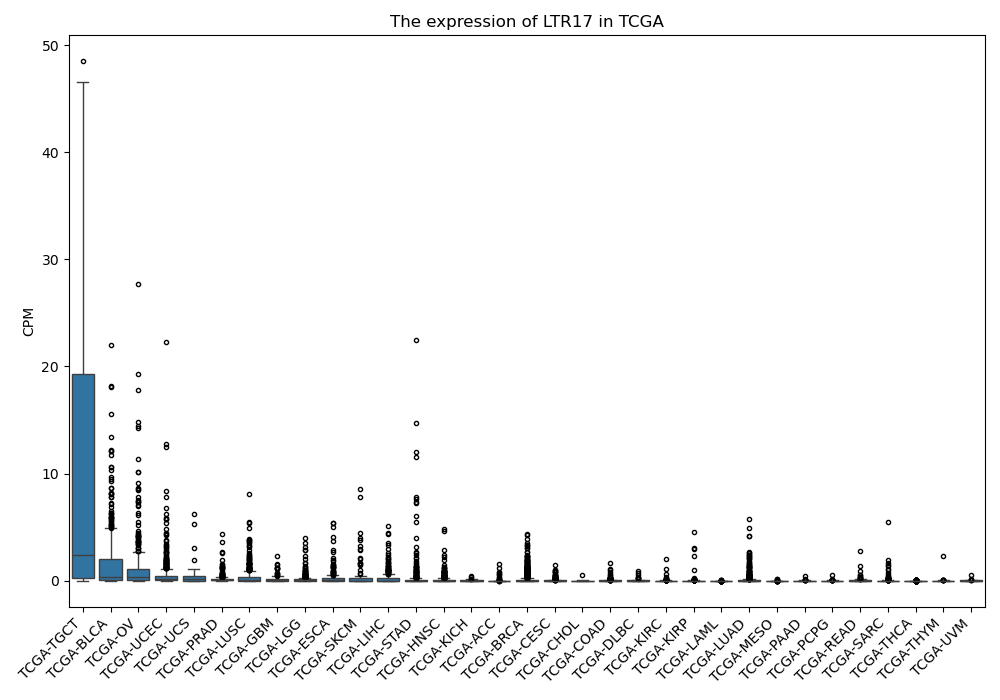
LTR17
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000427 |
|---|---|
| TE superfamily | ERV1 |
| TE class | LTR |
| Species | Catarrhini |
| Length | 780 |
| Kimura value | 4.64 |
| Tau index | 0.9806 |
| Description | LTR17 (Long Terminal Repeat) for HERV17 endogenous retrovirus |
| Comment | LTR17/HERV17 elements are flanked by 4 bp direct repeats. |
| Sequence |
TGAGAGACAGGACTAGCTGGATTTCCTAGGCCGACTAAGAATCCCTAAGCCTAGCTGGGAAGGTGACCGCATCCACCTTTAAACACGGGGCTTGCAACTTAGCTCACACCCGACCAATCAGGTAGTAAAGAGAGCTCACTAAAATGCTAATTAGGCAAAAACAGGAGGTAAAGAAATAGCCAATCATCTATCGCCTGAGAGCACAGCGGGAGGGACAATGATCGGGATATAAACCCAGGCATTCGAGCCGGCAACGGCNACCCCCTTTGGGTCCCCTCCCTTTGTATGGGAGCTCTGTTTTCACTCTATTAAATCTTGCAACTGCACTCTTCTGGTCCGTGTTTGTTACGGCTCGAGCTGAGCTTTCGCTCGCCGTCCACCACTGCTGTTTGCCGCCGTCGCAGACCCGCCGCTGACTTCCATCCCTCCGGATCCGGCAGGGTGTCCGCTGTGCTCCTGATCCAGCGAGGCGCCCATTGCCGCTCCCGATCGGGCTAAAGGCTTGCCATTGTTCCTGCACGGCTAAGTGCCCGGGTTCGTCCTAATCGAGCTGAACACTAGTCACTGGGTTCCACGGTTCTCTTCCGTGACCCACGGCTTCTAATAGAGCTATAACACTCACCGCATGGCCCAAGATTCCATTCCTTGGAATCCGTGAGGCCAAGAACCCCAGGTCAGAGAACACGAGGCTTGCCACCATCTTGGAAGCGGCCCGCCGCCATTTTGGAAGCGGCCCGCCACCATCTTGGGAGCTCTGGGAGCAAGGACCCCCCGGTAACA
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| LTR17 | ERF4 | 713 | 721 | + | 16.16 | CCGCCGCCA |
| LTR17 | ERF077 | 713 | 722 | + | 16.14 | CCGCCGCCAT |
| LTR17 | PLT1 | 644 | 659 | - | 16.12 | CTCACGGATTCCAAGG |
| LTR17 | ERF112 | 713 | 721 | + | 16.11 | CCGCCGCCA |
| LTR17 | MAZ | 273 | 280 | + | 16.08 | CCCCTCCC |
| LTR17 | REF6 | 293 | 301 | - | 15.93 | AAAACAGAG |
| LTR17 | POU1F1 | 141 | 154 | - | 15.69 | CTAATTAGCATTTT |
| LTR17 | ZIC1 | 202 | 215 | - | 15.64 | TCCCTCCCGCTGTG |
| LTR17 | ERF118 | 713 | 720 | + | 15.56 | CCGCCGCC |
| LTR17 | IG1 | 713 | 721 | + | 15.50 | CCGCCGCCA |
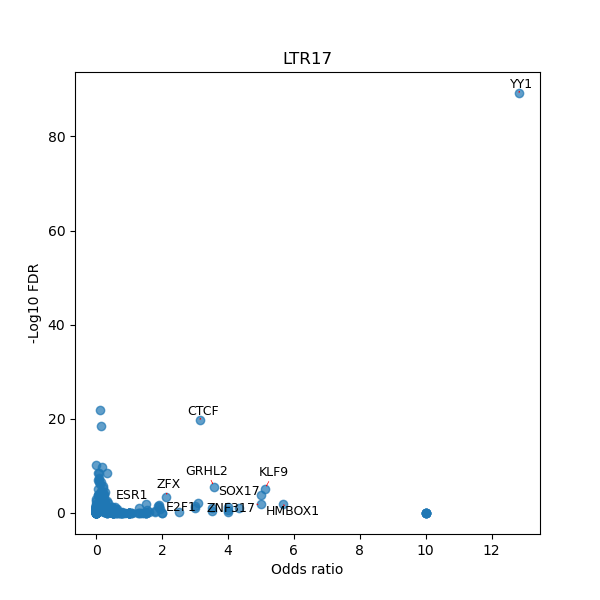
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.