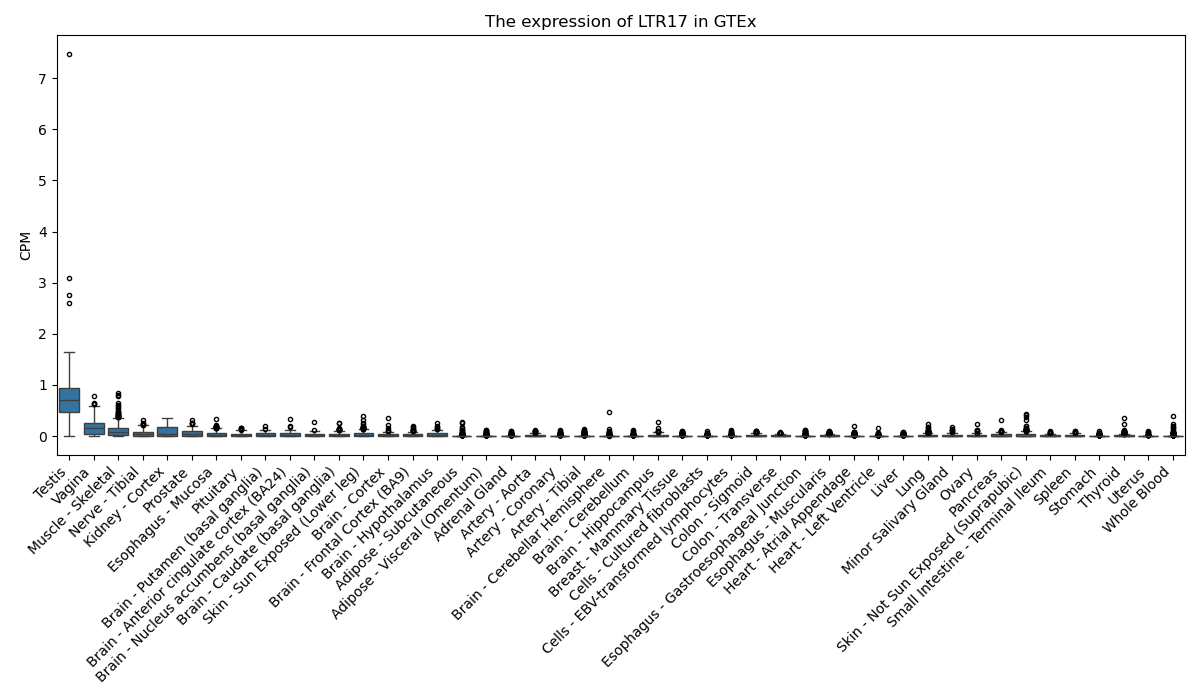
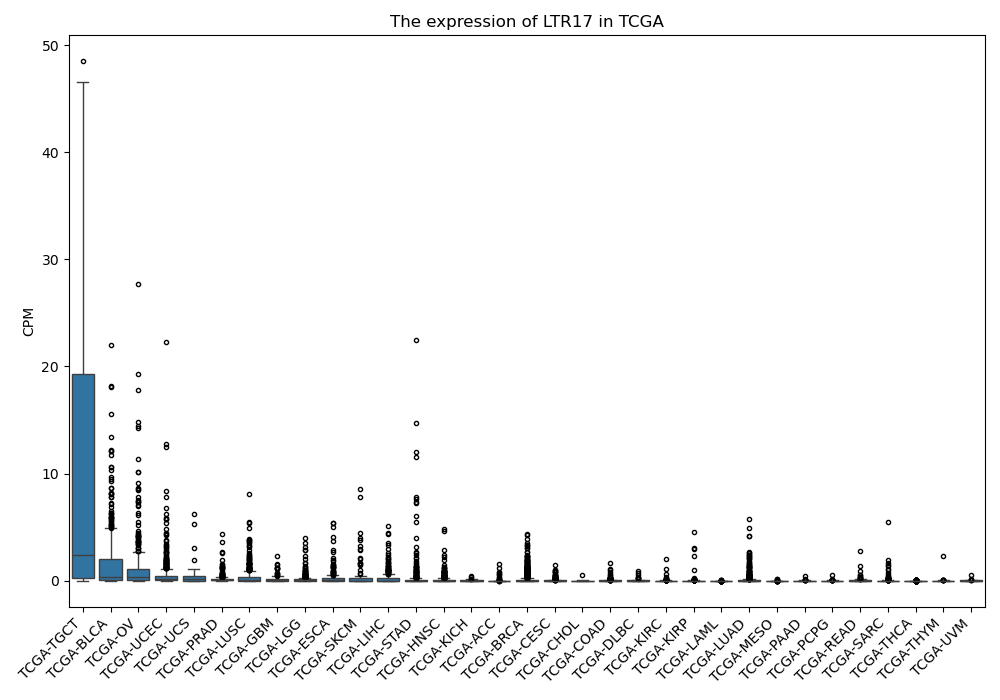
LTR17
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000427 |
|---|---|
| TE superfamily | ERV1 |
| TE class | LTR |
| Species | Catarrhini |
| Length | 780 |
| Kimura value | 4.64 |
| Tau index | 0.9806 |
| Description | LTR17 (Long Terminal Repeat) for HERV17 endogenous retrovirus |
| Comment | LTR17/HERV17 elements are flanked by 4 bp direct repeats. |
| Sequence |
TGAGAGACAGGACTAGCTGGATTTCCTAGGCCGACTAAGAATCCCTAAGCCTAGCTGGGAAGGTGACCGCATCCACCTTTAAACACGGGGCTTGCAACTTAGCTCACACCCGACCAATCAGGTAGTAAAGAGAGCTCACTAAAATGCTAATTAGGCAAAAACAGGAGGTAAAGAAATAGCCAATCATCTATCGCCTGAGAGCACAGCGGGAGGGACAATGATCGGGATATAAACCCAGGCATTCGAGCCGGCAACGGCNACCCCCTTTGGGTCCCCTCCCTTTGTATGGGAGCTCTGTTTTCACTCTATTAAATCTTGCAACTGCACTCTTCTGGTCCGTGTTTGTTACGGCTCGAGCTGAGCTTTCGCTCGCCGTCCACCACTGCTGTTTGCCGCCGTCGCAGACCCGCCGCTGACTTCCATCCCTCCGGATCCGGCAGGGTGTCCGCTGTGCTCCTGATCCAGCGAGGCGCCCATTGCCGCTCCCGATCGGGCTAAAGGCTTGCCATTGTTCCTGCACGGCTAAGTGCCCGGGTTCGTCCTAATCGAGCTGAACACTAGTCACTGGGTTCCACGGTTCTCTTCCGTGACCCACGGCTTCTAATAGAGCTATAACACTCACCGCATGGCCCAAGATTCCATTCCTTGGAATCCGTGAGGCCAAGAACCCCAGGTCAGAGAACACGAGGCTTGCCACCATCTTGGAAGCGGCCCGCCGCCATTTTGGAAGCGGCCCGCCACCATCTTGGGAGCTCTGGGAGCAAGGACCCCCCGGTAACA
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| LTR17 | RAP2-12 | 713 | 723 | - | 18.35 | AATGGCGGCGG |
| LTR17 | phol | 717 | 726 | + | 17.57 | CGCCATTTTG |
| LTR17 | ZFP42 | 714 | 726 | - | 17.54 | CAAAATGGCGGCG |
| LTR17 | EREB138 | 713 | 722 | + | 17.53 | CCGCCGCCAT |
| LTR17 | ERF105 | 713 | 722 | - | 16.95 | ATGGCGGCGG |
| LTR17 | nfya-1 | 112 | 121 | + | 16.78 | GACCAATCAG |
| LTR17 | YY1-2 | 716 | 728 | - | 16.62 | TCCAAAATGGCGG |
| LTR17 | AIL6 | 644 | 658 | + | 16.61 | CCTTGGAATCCGTGA |
| LTR17 | ERF082 | 713 | 721 | + | 16.26 | CCGCCGCCA |
| LTR17 | Sox7 | 506 | 515 | - | 16.17 | GGAACAATGG |
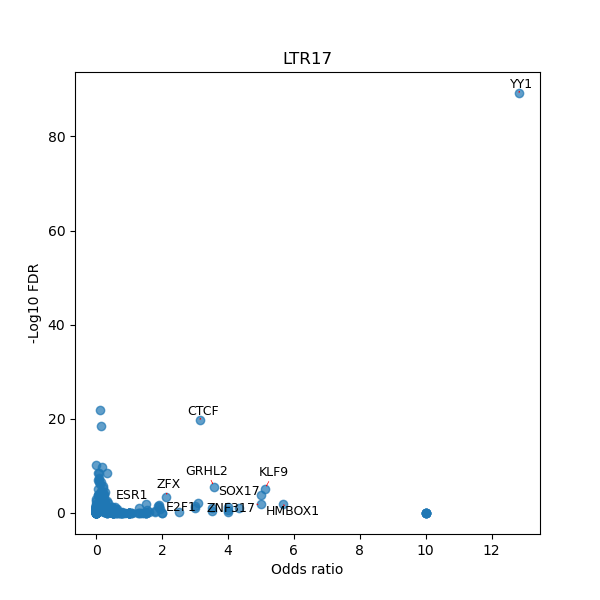
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.