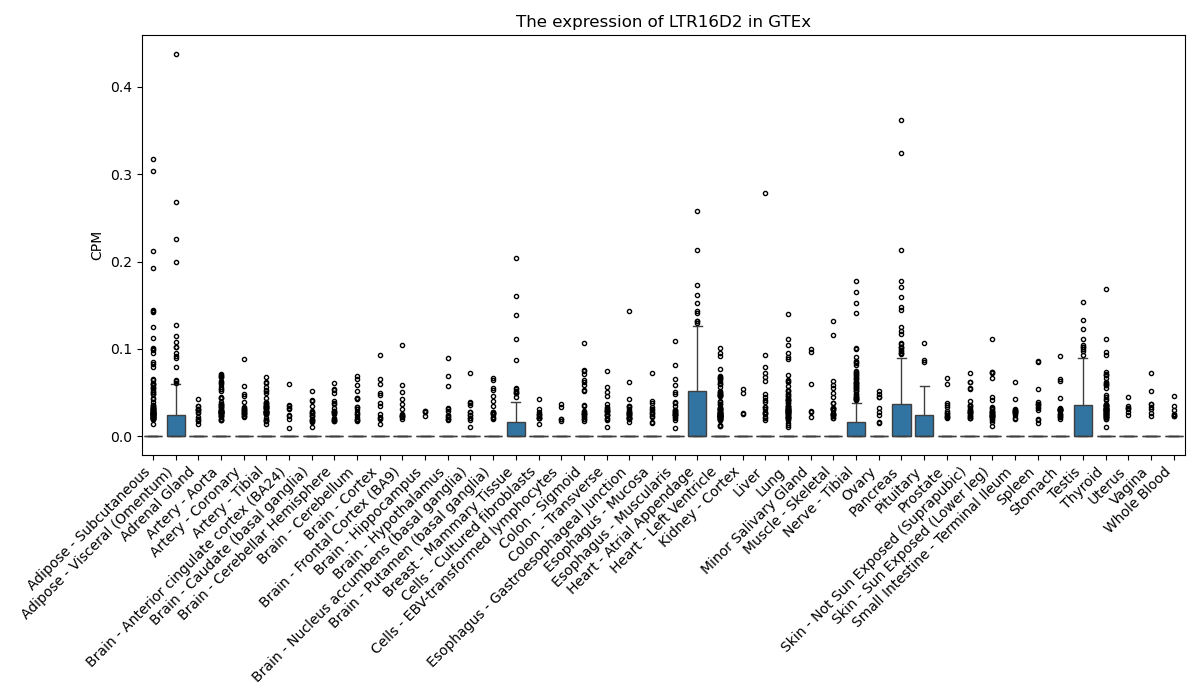
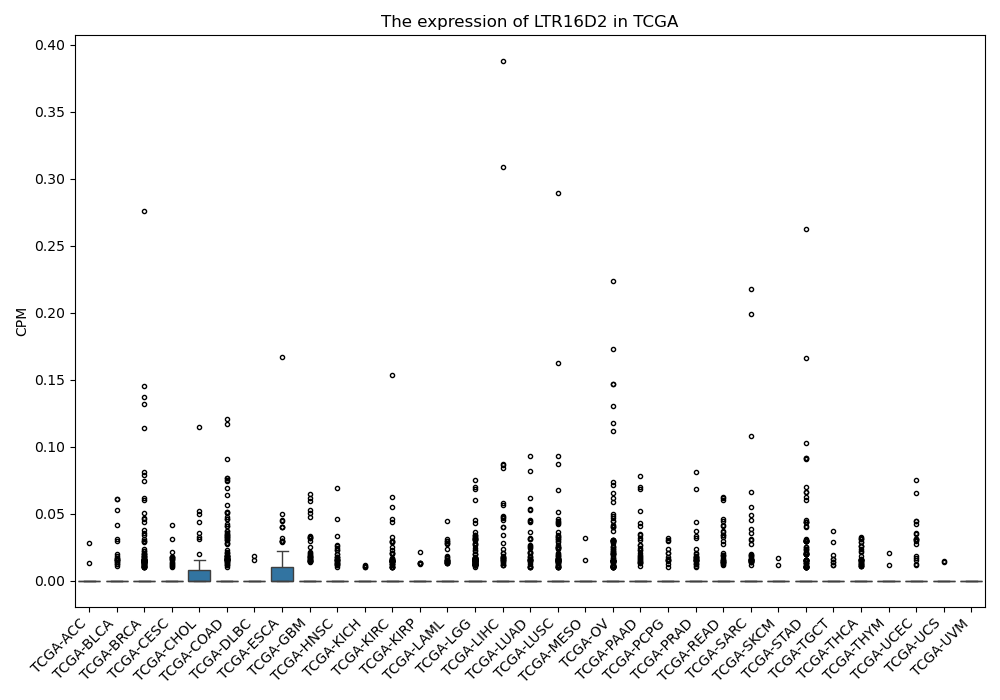
LTR16D2
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000424 |
|---|---|
| TE superfamily | ERV3 |
| TE class | LTR |
| Species | Eutheria |
| Length | 572 |
| Kimura value | 29.63 |
| Tau index | 0.0000 |
| Description | LTR16D2 (Long Terminal Repeat) for ERVL endogenous retrovirus |
| Comment | - |
| Sequence |
TGTATTGGACACAAACCTTGTGCGCCTTCTCAGATTCCCTCGACCGCCTCCTTCTTTCTCTCTCTTGGCCGGCGCCCCACCCGCGCCGGGTCGCCCCTCCACTTTCGGACTTGGATGAAACGCCACACCGCGGGGCATGGGATCTGGCTTCCTGAGCGGCCGCCGAAGGGGCCGGATGACGCAACCCGGAAGTGCAGGGGAGTTAGCTCCCCGTGGGGTGAACCTTGACCAATGGGAAACGGGAGACGGGAGGGAGCCGGGCAGATAAATTCCCCCTCCTTTCTCTCTTCCGTGGACTACTCCGAGGTGCGGTTCCTCCTTGCAACCCTTCCGGAGAAGTCCCGCGTGCCGAGCGAACACGCCTGCTGAGCGACCTGCTGTGTCTCTTCGCGGCTCGTTGTGAAGCGGTAGCCAGCGCGGTAACGCATCGCATCGCATTGCTTCGCATCTTTCCTTGCCTCACTTCCCTTTTTCCTCACCCTCGCCGCCCTGGGCTTGCACCTCCCAAATAAAGTGTTAGCACCTTAATCCTTGCCTCAGGCTCTGCTTTCTAGAGGACCCGGGCTAAGACA
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| LTR16D2 | Trl | 281 | 289 | - | 14.34 | AAGAGAGAA |
| LTR16D2 | FOXJ2::ELF1 | 183 | 193 | + | 14.33 | AACCCGGAAGT |
| LTR16D2 | ETV7 | 186 | 194 | + | 14.31 | CCGGAAGTG |
| LTR16D2 | ZNF257 | 452 | 461 | - | 14.22 | GAGGCAAGGA |
| LTR16D2 | ZNF257 | 529 | 538 | - | 14.22 | GAGGCAAGGA |
| LTR16D2 | eor-1 | 53 | 65 | - | 14.11 | AGAGAGAGAAAGA |
| LTR16D2 | MAZ | 273 | 280 | + | 14.06 | CCCCTCCT |
| LTR16D2 | Ets65A | 186 | 193 | + | 13.95 | CCGGAAGT |
| LTR16D2 | BPC6 | 46 | 66 | + | 13.93 | GCCTCCTTCTTTCTCTCTCTT |
| LTR16D2 | Zic1::Zic2 | 362 | 368 | - | 13.93 | CAGCAGG |
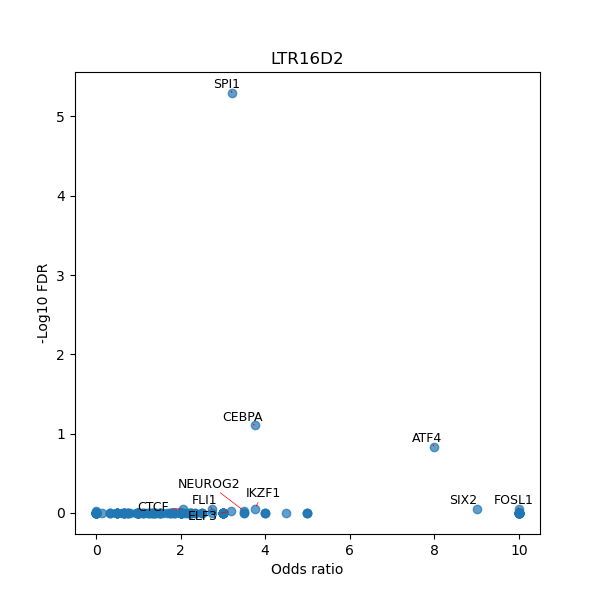
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.