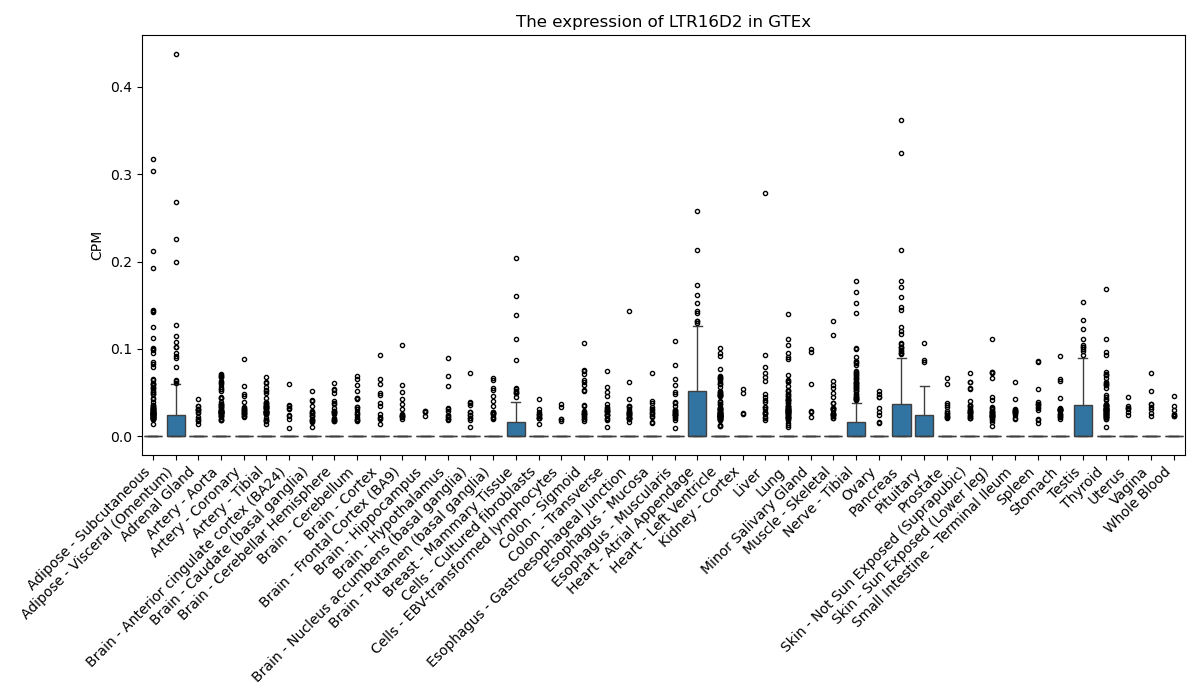
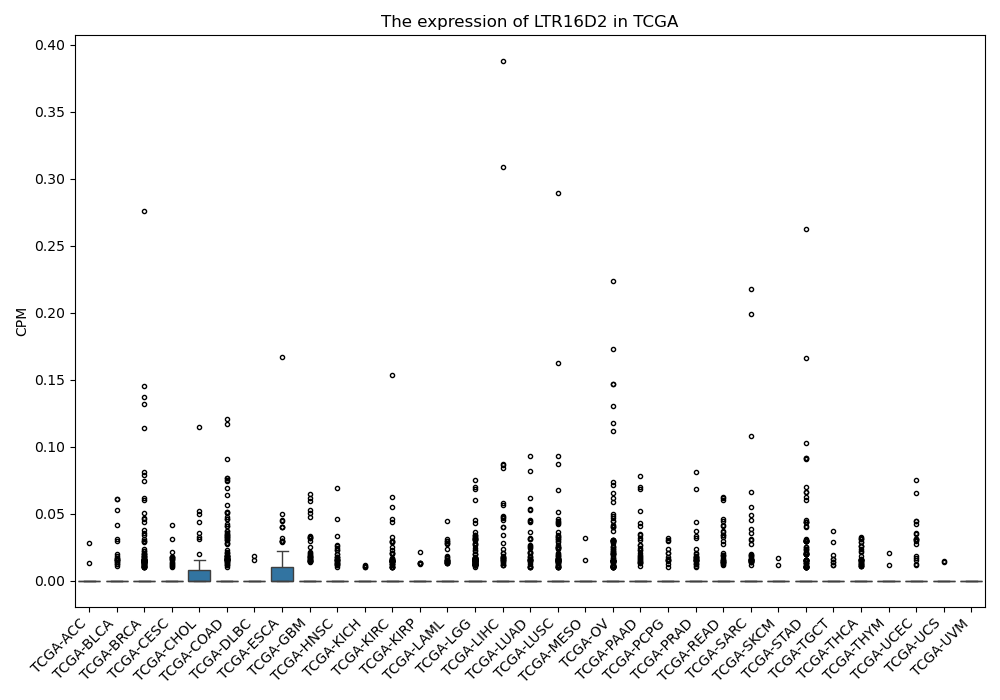
LTR16D2
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000424 |
|---|---|
| TE superfamily | ERV3 |
| TE class | LTR |
| Species | Eutheria |
| Length | 572 |
| Kimura value | 29.63 |
| Tau index | 0.0000 |
| Description | LTR16D2 (Long Terminal Repeat) for ERVL endogenous retrovirus |
| Comment | - |
| Sequence |
TGTATTGGACACAAACCTTGTGCGCCTTCTCAGATTCCCTCGACCGCCTCCTTCTTTCTCTCTCTTGGCCGGCGCCCCACCCGCGCCGGGTCGCCCCTCCACTTTCGGACTTGGATGAAACGCCACACCGCGGGGCATGGGATCTGGCTTCCTGAGCGGCCGCCGAAGGGGCCGGATGACGCAACCCGGAAGTGCAGGGGAGTTAGCTCCCCGTGGGGTGAACCTTGACCAATGGGAAACGGGAGACGGGAGGGAGCCGGGCAGATAAATTCCCCCTCCTTTCTCTCTTCCGTGGACTACTCCGAGGTGCGGTTCCTCCTTGCAACCCTTCCGGAGAAGTCCCGCGTGCCGAGCGAACACGCCTGCTGAGCGACCTGCTGTGTCTCTTCGCGGCTCGTTGTGAAGCGGTAGCCAGCGCGGTAACGCATCGCATCGCATTGCTTCGCATCTTTCCTTGCCTCACTTCCCTTTTTCCTCACCCTCGCCGCCCTGGGCTTGCACCTCCCAAATAAAGTGTTAGCACCTTAATCCTTGCCTCAGGCTCTGCTTTCTAGAGGACCCGGGCTAAGACA
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| LTR16D2 | BPC1 | 43 | 66 | - | 15.32 | AAGAGAGAGAAAGAAGGAGGCGGT |
| LTR16D2 | Ets21C | 186 | 193 | + | 15.09 | CCGGAAGT |
| LTR16D2 | ETV6 | 186 | 194 | + | 14.92 | CCGGAAGTG |
| LTR16D2 | SPDEF | 184 | 193 | + | 14.69 | ACCCGGAAGT |
| LTR16D2 | KLF1 | 75 | 82 | - | 14.64 | GGGTGGGG |
| LTR16D2 | Mad | 75 | 89 | - | 14.63 | CCGGCGCGGGTGGGG |
| LTR16D2 | Zbtb2 | 185 | 194 | + | 14.53 | CCCGGAAGTG |
| LTR16D2 | TFAP2E | 534 | 542 | + | 14.51 | GCCTCAGGC |
| LTR16D2 | TFAP2E | 534 | 542 | - | 14.47 | GCCTGAGGC |
| LTR16D2 | Stat6 | 329 | 338 | + | 14.45 | TTCCGGAGAA |
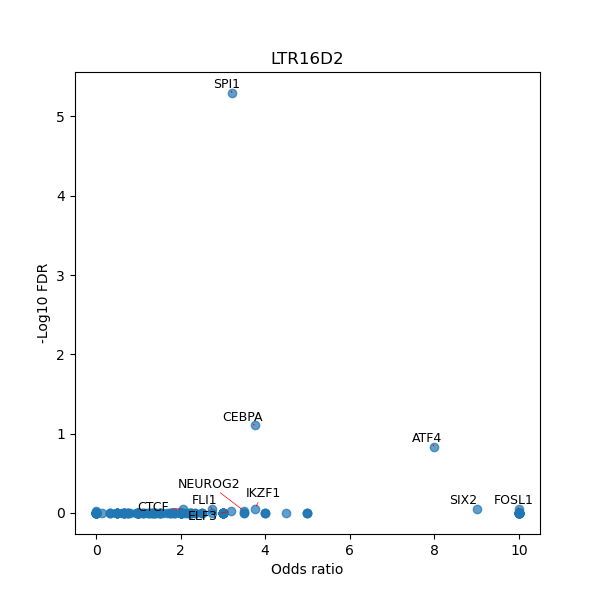
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.