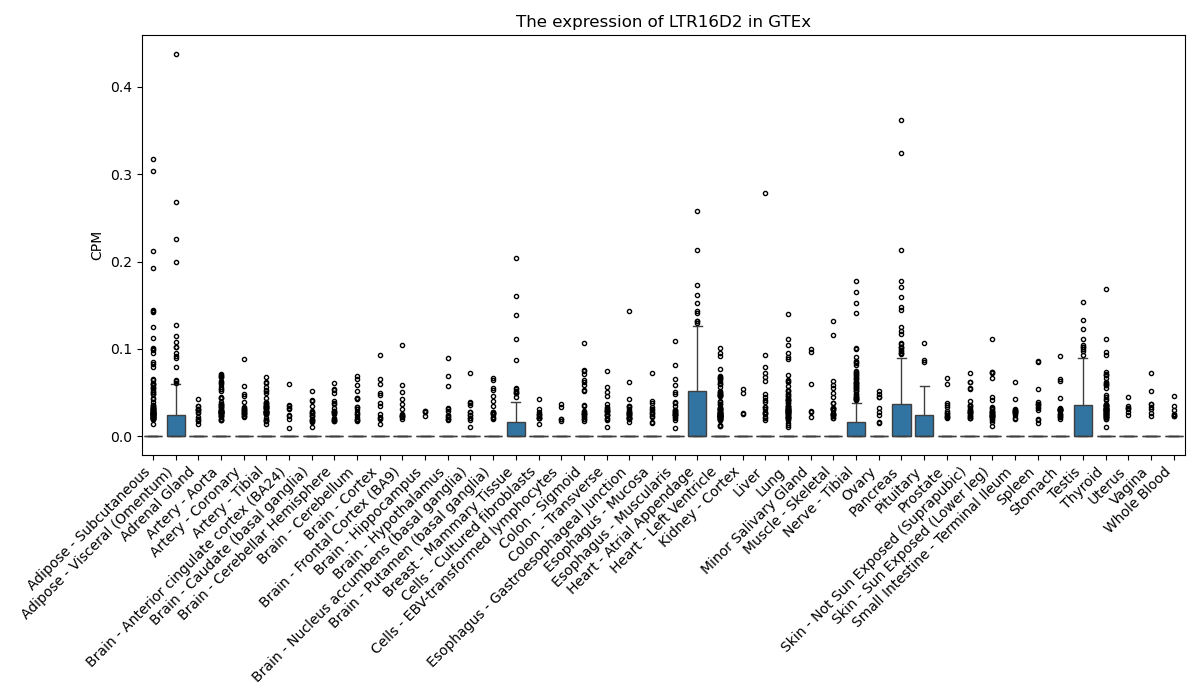
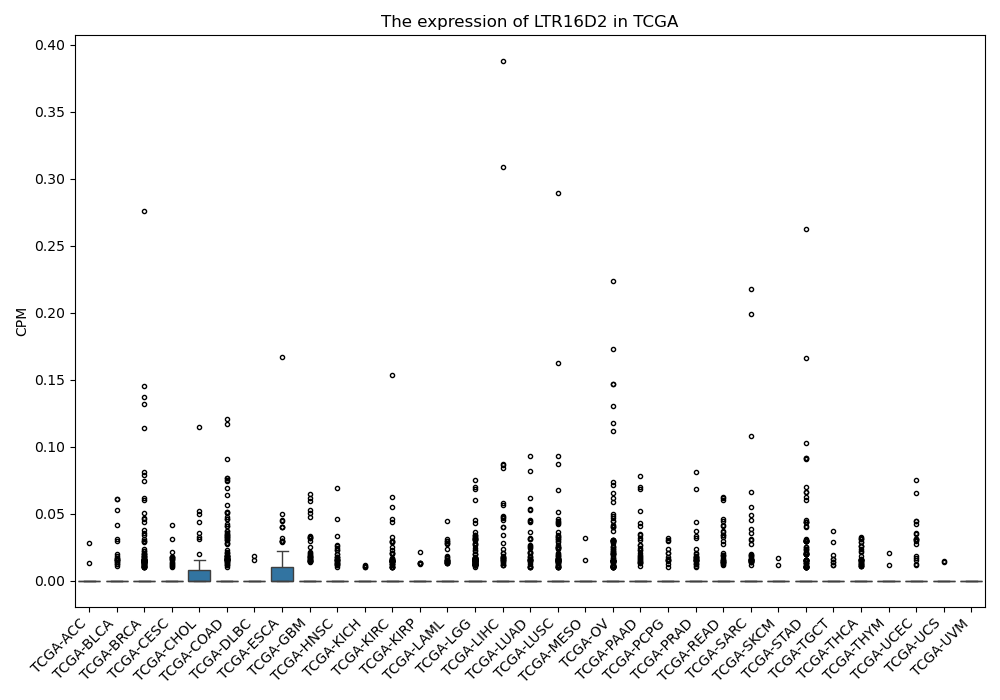
LTR16D2
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000424 |
|---|---|
| TE superfamily | ERV3 |
| TE class | LTR |
| Species | Eutheria |
| Length | 572 |
| Kimura value | 29.63 |
| Tau index | 0.0000 |
| Description | LTR16D2 (Long Terminal Repeat) for ERVL endogenous retrovirus |
| Comment | - |
| Sequence |
TGTATTGGACACAAACCTTGTGCGCCTTCTCAGATTCCCTCGACCGCCTCCTTCTTTCTCTCTCTTGGCCGGCGCCCCACCCGCGCCGGGTCGCCCCTCCACTTTCGGACTTGGATGAAACGCCACACCGCGGGGCATGGGATCTGGCTTCCTGAGCGGCCGCCGAAGGGGCCGGATGACGCAACCCGGAAGTGCAGGGGAGTTAGCTCCCCGTGGGGTGAACCTTGACCAATGGGAAACGGGAGACGGGAGGGAGCCGGGCAGATAAATTCCCCCTCCTTTCTCTCTTCCGTGGACTACTCCGAGGTGCGGTTCCTCCTTGCAACCCTTCCGGAGAAGTCCCGCGTGCCGAGCGAACACGCCTGCTGAGCGACCTGCTGTGTCTCTTCGCGGCTCGTTGTGAAGCGGTAGCCAGCGCGGTAACGCATCGCATCGCATTGCTTCGCATCTTTCCTTGCCTCACTTCCCTTTTTCCTCACCCTCGCCGCCCTGGGCTTGCACCTCCCAAATAAAGTGTTAGCACCTTAATCCTTGCCTCAGGCTCTGCTTTCTAGAGGACCCGGGCTAAGACA
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| LTR16D2 | ARF10 | 238 | 248 | + | 15.85 | AACGGGAGACG |
| LTR16D2 | ZKSCAN5 | 459 | 467 | - | 15.83 | GGAAGTGAG |
| LTR16D2 | PATZ1 | 74 | 84 | - | 15.64 | GCGGGTGGGGC |
| LTR16D2 | Trl | 58 | 66 | - | 15.63 | AAGAGAGAG |
| LTR16D2 | TfAP-2 | 534 | 542 | - | 15.56 | GCCTGAGGC |
| LTR16D2 | TFAP2B | 534 | 542 | - | 15.56 | GCCTGAGGC |
| LTR16D2 | TfAP-2 | 534 | 542 | + | 15.55 | GCCTCAGGC |
| LTR16D2 | KLF4 | 75 | 82 | + | 15.48 | CCCCACCC |
| LTR16D2 | E(spl)mbeta-HLH | 343 | 350 | + | 15.34 | CGCGTGCC |
| LTR16D2 | Zic2 | 362 | 370 | - | 15.33 | CTCAGCAGG |
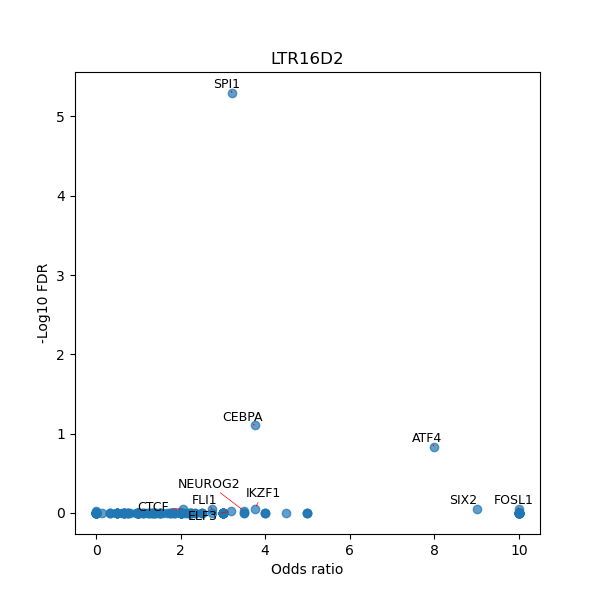
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.