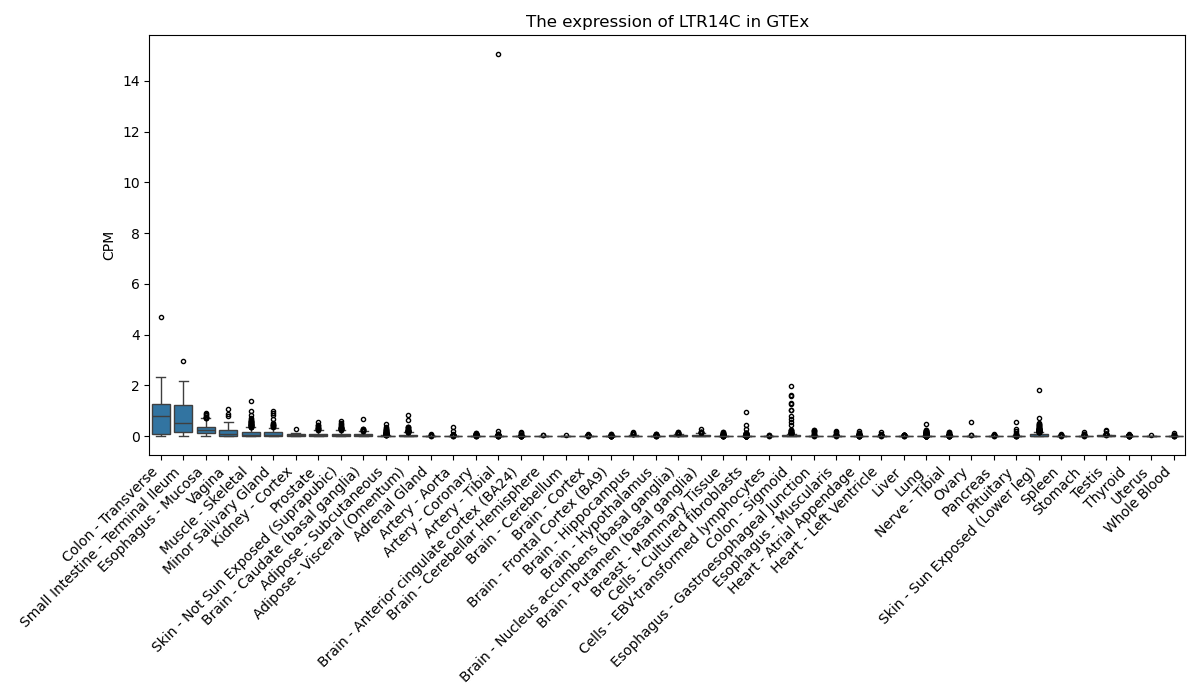
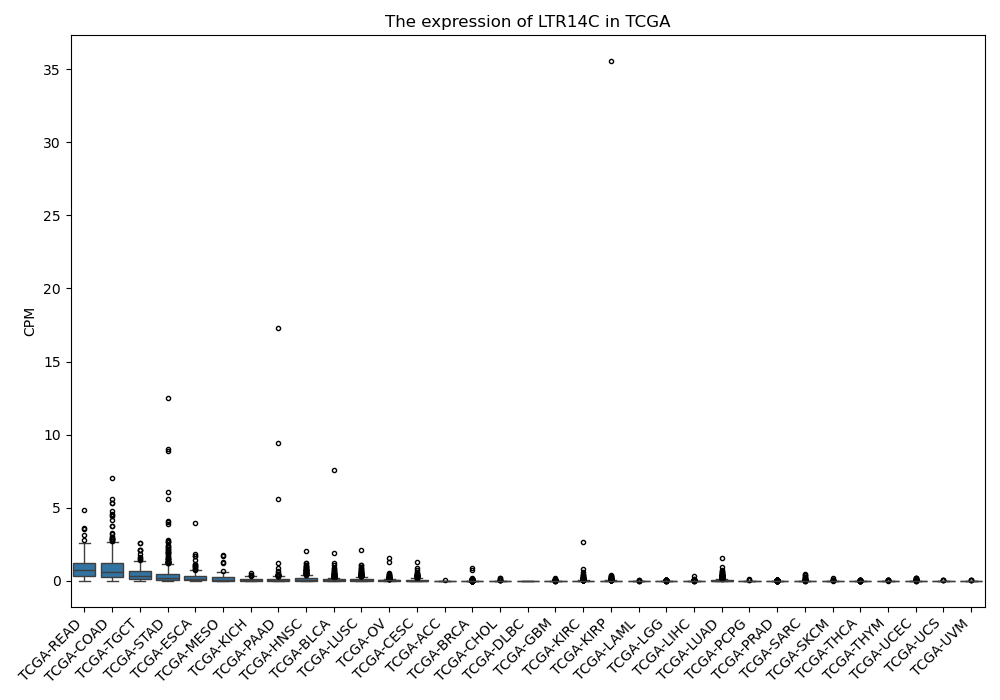
LTR14C
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000412 |
|---|---|
| TE superfamily | ERV2 |
| TE class | LTR |
| Species | Catarrhini |
| Length | 587 |
| Kimura value | 5.32 |
| Tau index | 0.9690 |
| Description | LTR14C (Long Terminal Repeat) for HERVK14C endogenous retrovirus |
| Comment | Copies present at orthologous sites in old world monkeys, but absent from new world monkeys. |
| Sequence |
TGTGGGAAAGAGAGTTTCTGGGGTGCCAGNTGAGTTGGTCTCCCCTGTGTGAGACACCCATGGGGAGCCATGGGCGGCCTCTGAGGAGAAAAGTCTCCTTATTGCCTTCATGTCTTTATGCCCCGAGAGCATAACCGCTCAGCGGCATTCCACAGGTTGCTCAGGGAGATAACACTCCCTTGAAGCAGTGGAGTATAATCAAACATCTTGGCTCCTCCTGAAACCCGCTCCCACCCGTTTCAGTCCCGATAAGTTAAAGATCTTAAGTAGTTTAGACACACGCCTTTGCTCAAGGAAATTCACAGAAACCGCCACTGCTATACATCTTATCGAATGACTCACGAGTTCTCCTTCACTGATTAATCCTTTTCCTCATCCCTTCCTCCCCCTCCCATCTGCCCTAAGAACAAAGAGCTTGTAAACCAATAAATTGGGCGGAGCCCGAGAGCTCTGGGCCGTGAGCAAGCCTCCGACGCTCCGGTCCCCTGGACCCGCCTTTTAAACGCTTATTCTGTCTCTTTCTAACTCCTTTGTCTCCGCCGGACTCGGGGTACCCGCCGGGTGGTGTGGGGCTGGTTTCCCCAACA
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| LTR14C | FOS::JUN | 334 | 342 | - | 13.93 | GTGAGTCAT |
| LTR14C | FOS | 334 | 341 | - | 13.87 | TGAGTCAT |
| LTR14C | SP4 | 386 | 394 | - | 13.83 | TGGGAGGGG |
| LTR14C | Sox6 | 528 | 537 | + | 13.83 | CCTTTGTCTC |
| LTR14C | ZNF263 | 381 | 387 | - | 13.83 | GGGAGGA |
| LTR14C | PRDM9 | 372 | 391 | - | 13.81 | GAGGGGGAGGAAGGGATGAG |
| LTR14C | BACH2 | 333 | 343 | - | 13.80 | CGTGAGTCATT |
| LTR14C | Sox14 | 405 | 415 | - | 13.79 | GCTCTTTGTTC |
| LTR14C | fos-1 | 335 | 341 | + | 13.76 | TGACTCA |
| LTR14C | Spps | 431 | 441 | - | 13.70 | GCTCCGCCCAA |
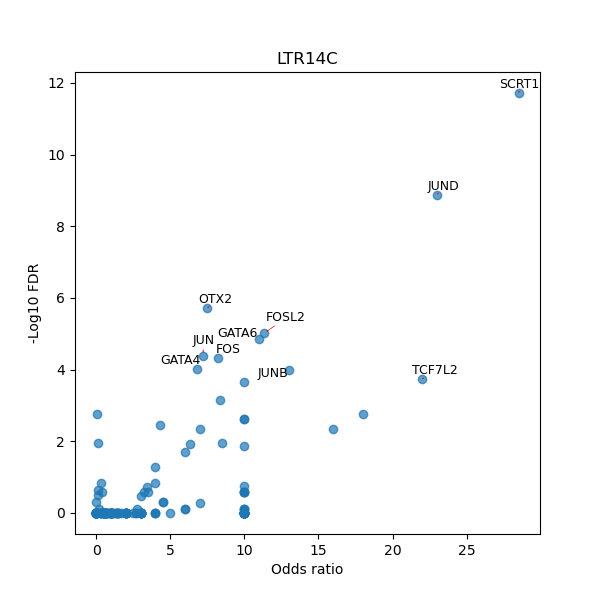
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.