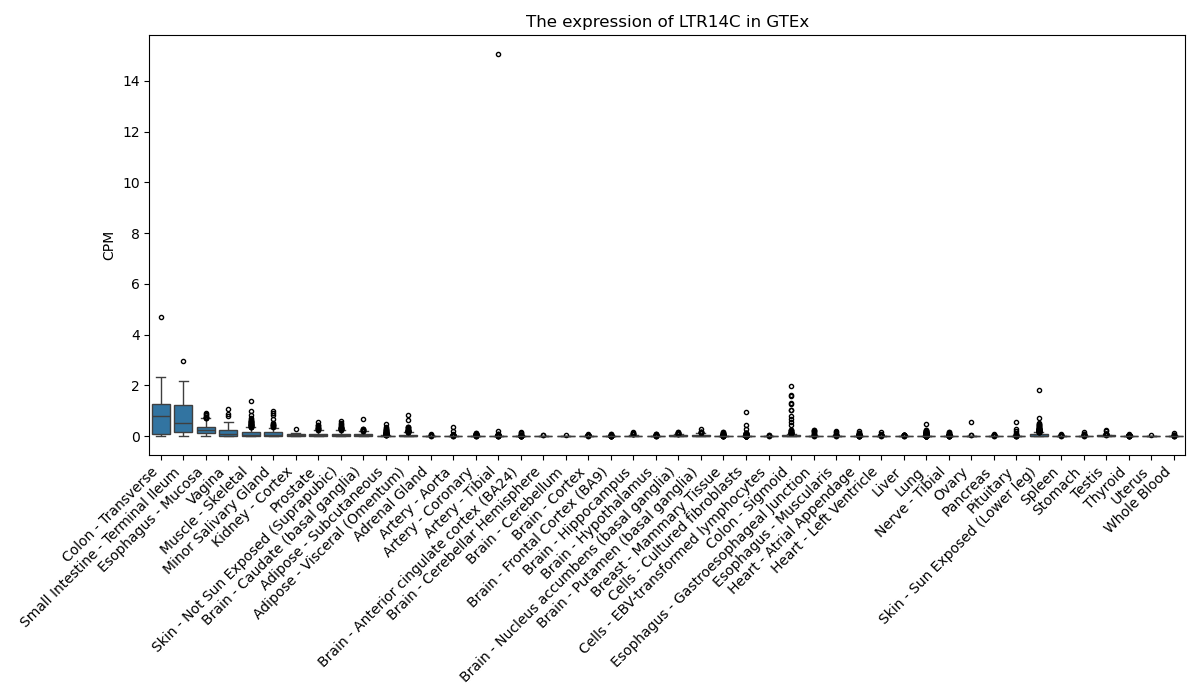
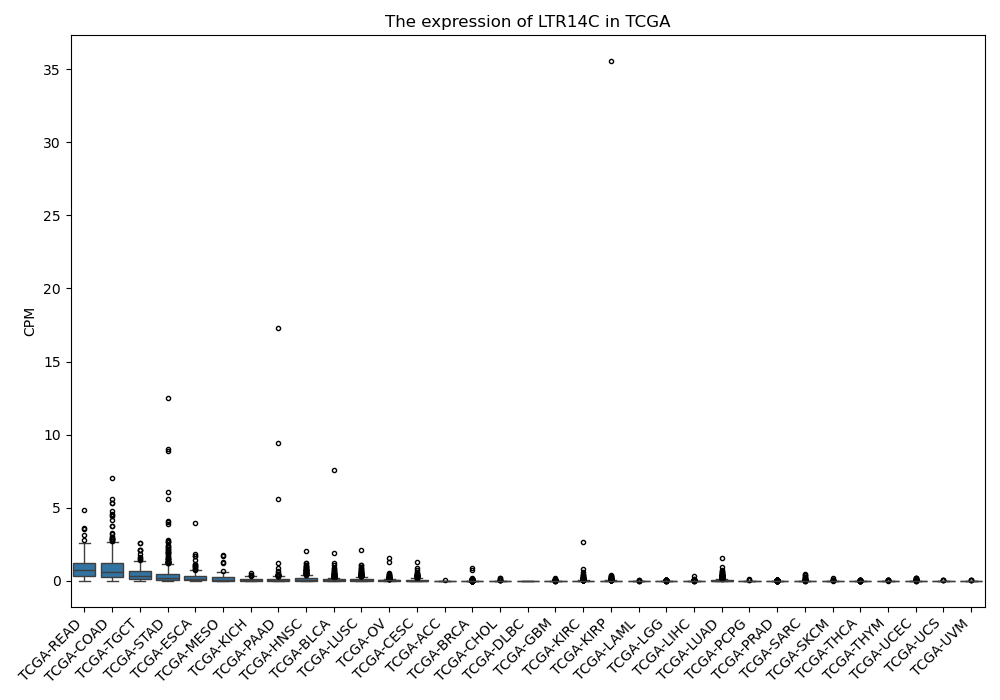
LTR14C
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000412 |
|---|---|
| TE superfamily | ERV2 |
| TE class | LTR |
| Species | Catarrhini |
| Length | 587 |
| Kimura value | 5.32 |
| Tau index | 0.9690 |
| Description | LTR14C (Long Terminal Repeat) for HERVK14C endogenous retrovirus |
| Comment | Copies present at orthologous sites in old world monkeys, but absent from new world monkeys. |
| Sequence |
TGTGGGAAAGAGAGTTTCTGGGGTGCCAGNTGAGTTGGTCTCCCCTGTGTGAGACACCCATGGGGAGCCATGGGCGGCCTCTGAGGAGAAAAGTCTCCTTATTGCCTTCATGTCTTTATGCCCCGAGAGCATAACCGCTCAGCGGCATTCCACAGGTTGCTCAGGGAGATAACACTCCCTTGAAGCAGTGGAGTATAATCAAACATCTTGGCTCCTCCTGAAACCCGCTCCCACCCGTTTCAGTCCCGATAAGTTAAAGATCTTAAGTAGTTTAGACACACGCCTTTGCTCAAGGAAATTCACAGAAACCGCCACTGCTATACATCTTATCGAATGACTCACGAGTTCTCCTTCACTGATTAATCCTTTTCCTCATCCCTTCCTCCCCCTCCCATCTGCCCTAAGAACAAAGAGCTTGTAAACCAATAAATTGGGCGGAGCCCGAGAGCTCTGGGCCGTGAGCAAGCCTCCGACGCTCCGGTCCCCTGGACCCGCCTTTTAAACGCTTATTCTGTCTCTTTCTAACTCCTTTGTCTCCGCCGGACTCGGGGTACCCGCCGGGTGGTGTGGGGCTGGTTTCCCCAACA
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| LTR14C | ZAP1 | 553 | 567 | + | -3.36 | ACCCGCCGGGTGGTG |
| LTR14C | EWSR1-FLI1 | 366 | 383 | - | -5.26 | GGAAGGGATGAGGAAAAG |
| LTR14C | EWSR1-FLI1 | 374 | 391 | - | -5.54 | GAGGGGGAGGAAGGGATG |
| LTR14C | BPC5 | 504 | 533 | - | -31.63 | CAAAGGAGTTAGAAAGAGACAGAATAAGCG |
| LTR14C | BPC5 | 502 | 531 | - | -42.32 | AAGGAGTTAGAAAGAGACAGAATAAGCGTT |
| LTR14C | BPC5 | 506 | 535 | - | -46.34 | GACAAAGGAGTTAGAAAGAGACAGAATAAG |
| LTR14C | BPC5 | 494 | 523 | - | -48.05 | AGAAAGAGACAGAATAAGCGTTTAAAAGGC |
| LTR14C | BPC5 | 498 | 527 | - | -48.16 | AGTTAGAAAGAGACAGAATAAGCGTTTAAA |
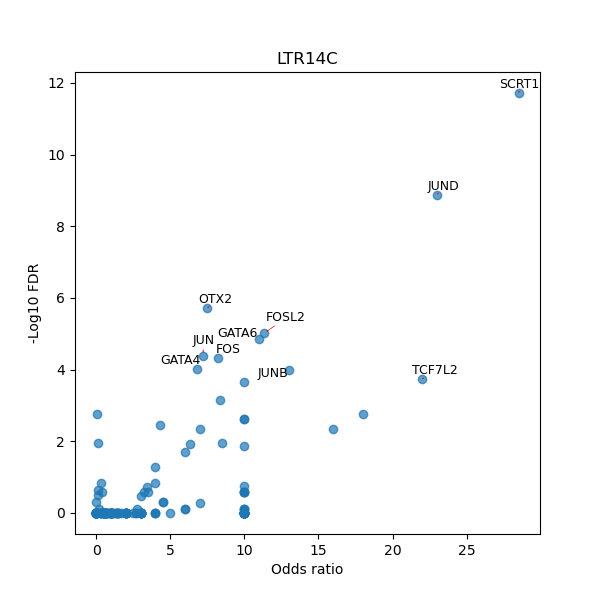
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.