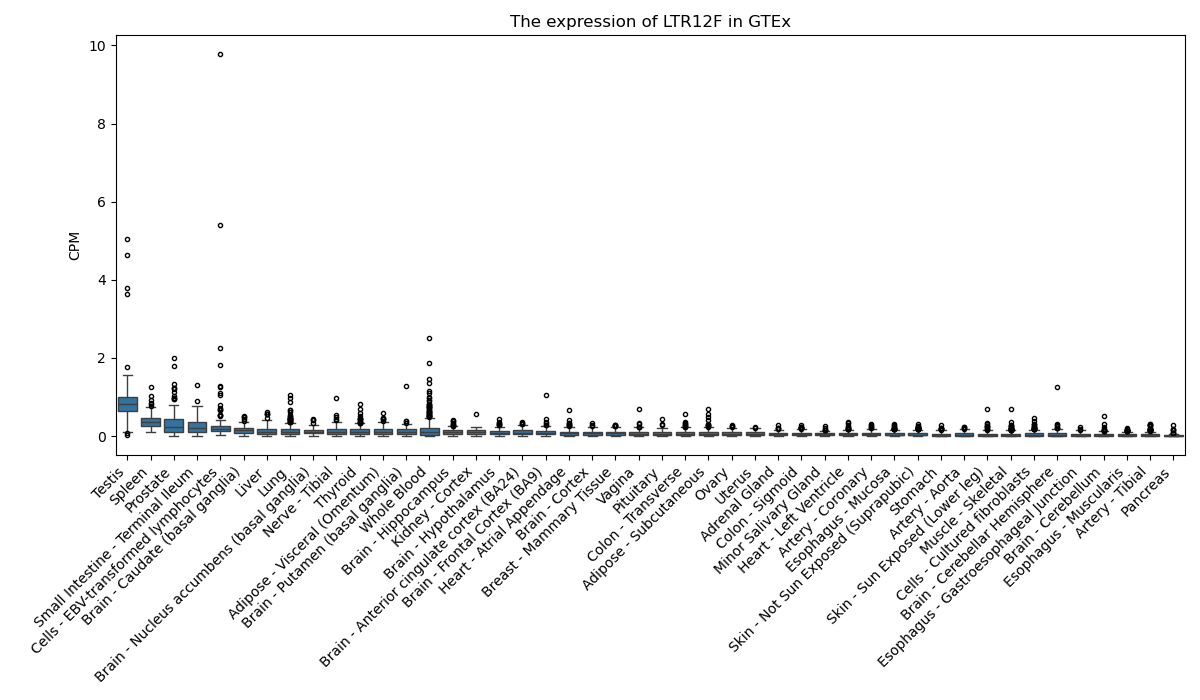
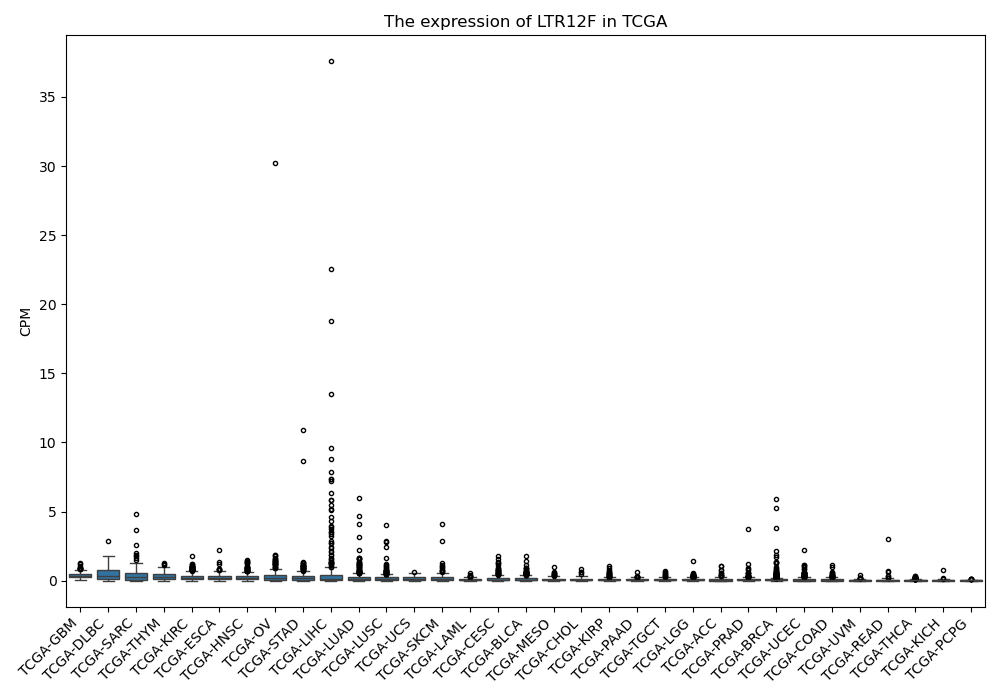
LTR12F
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000405 |
|---|---|
| TE superfamily | ERV1 |
| TE class | LTR |
| Species | Catarrhini |
| Length | 519 |
| Kimura value | 6.71 |
| Tau index | 0.9044 |
| Description | Long terminal repeat of class I endogenous retrovirus HERV9 |
| Comment | Positions 73-207 of the consensus contains 5 copies of a 23-bp-period tandem repeat interrupted by a 15 bp sequence. The number of units at any locus is variable. Matches limited to pos. ~334 to 411 represent retroposed copies of the region of the LTR between the transcription initiation and termination signals (named R as it Repeats at the ends of the transcript). They are followed by a poly A tail and were likely introduced by the LINE1 machinery. They could be the result of R-R recombination of a LINE1-retroposed full transcripts, though no full processed psuedogenes of HERV9 with LTR12F LTRs survived. Slight target site duplication preference for MTAK. |
| Sequence |
TGAGAGGTGAAGCCAGCTGGACTTCCTGGGTCGAGTGGGGACTTGGAGAACTTTTCTGTCTTACAAGAGGATTGTAAAATGCACCAATCAGCGCTCTGTAAAAACGCACCAATCAGCGCTCTGTAGCTAGCAAGAGGATTGTAAAATGCACCAATCAGCGCTCTGTAAAACGCACCAATCAGCGCTCTGTAAAACGCACCAATCAGCAGGATTCTAAAAGTAGCCAATCGCGGGGAGGATTGAAAAAAGGGCACTCTGATAGGACAGAAACGGAACATGGGAGGGGACAAATAAGGGAATAAAAGCTGGCCACCCCAGCCAGCAGCGGCAACCCGCTCGGGTCCCCTTCCACGCTGTGGAAGCTTTGTTCTTTCGCTCTTCACAATAAACCTTGCTGCCGCTCACTCTTTGGGTCCGTGCCATCTTTAAGAGCTGTAACACTCACCGCGAAGGTCCGCGGCTTCATTCTTGAAGTCAGCGAGACCACGAACCCACCGGNAGGAACCAACTCCGGACACA
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| LTR12F | tin | 30 | 38 | + | 13.94 | GTCGAGTGG |
| LTR12F | Zic1::Zic2 | 204 | 210 | + | 13.93 | CAGCAGG |
| LTR12F | BZIP30 | 12 | 20 | + | 13.87 | GCCAGCTGG |
| LTR12F | SP4 | 278 | 286 | + | 13.83 | TGGGAGGGG |
| LTR12F | ZNF263 | 233 | 239 | + | 13.83 | GGGAGGA |
| LTR12F | Zic3 | 204 | 210 | + | 13.70 | CAGCAGG |
| LTR12F | ETV1 | 21 | 29 | - | 13.70 | CCAGGAAGT |
| LTR12F | Ikzf3 | 20 | 28 | - | 13.67 | CAGGAAGTC |
| LTR12F | VIP1 | 11 | 20 | + | 13.64 | AGCCAGCTGG |
| LTR12F | ZNF175 | 21 | 29 | - | 13.64 | CCAGGAAGT |
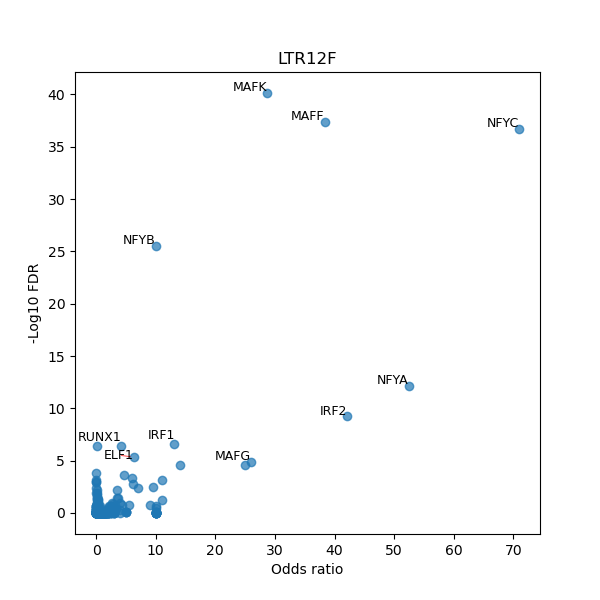
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.