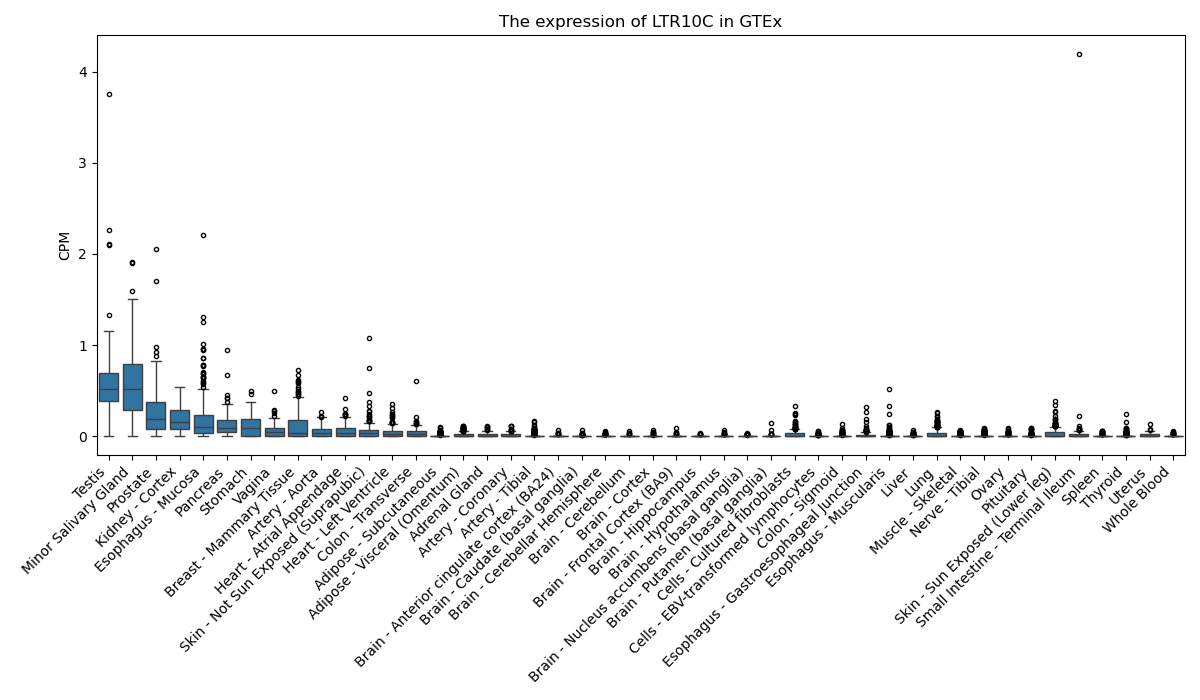
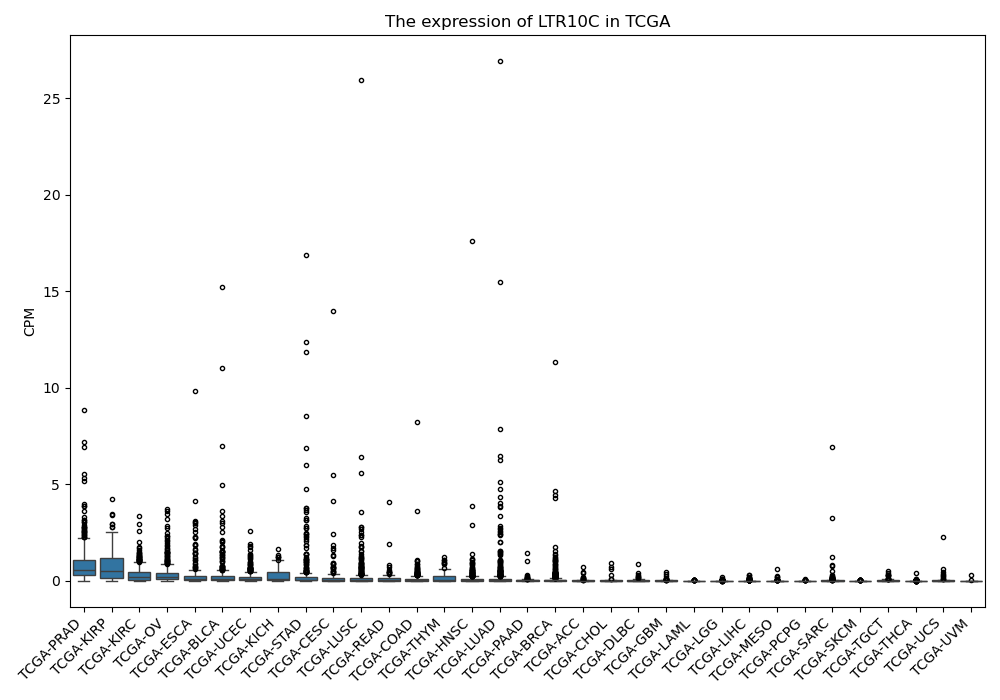
LTR10C
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000394 |
|---|---|
| TE superfamily | ERV1 |
| TE class | LTR |
| Species | Catarrhini |
| Length | 591 |
| Kimura value | 4.93 |
| Tau index | 0.9412 |
| Description | LTR10C (Long Terminal Repeat) for HERVI endogenous retrovirus |
| Comment | LTR10C is a long terminal repeat that appears to be associated with HERVI. |
| Sequence |
TGTAAAAAGTAAAGTAGAGGTTCCTCTTCAAAGACTTTCCTCCCCGTCTAATTAGGAATAAATAGTAACTTCTCTTAGAAGCAAAATTTATTCAAAGACCTGTGCTAACATTCTTAAATATCTGCTAGCCGTAATAAAGAAATCAATGTACTTTATGTTCTTAGCTCCCACAATTTAGCCTAAATATTTGCCCTGGCATGCTTATACTGGTCCAAGCAAGCATTAGGTCATAGCCTGTTCCTCTTCCTTATTTGAAGGTGTTTTTACCTTTCTCAGCATTCCACAAGTTACTTCCTCCTTCCTTTGTTCTCCTCTGCCTTTGCCTCTTTTAAAAAGTTCTAAGTTGCTAGCCAATCGGGACAAATACAGAATGTGAGGTCCCGTTCCAGCCAATGGAAACCGGACACAGCAGTAGGGTGGACGCGTCAGGTTATAAATGACCCTGTCTCCTTTGTTCGGTGTACTCTCGTGGCAAAACTGCTGGCGAGTGTACCCTTTCTGCAGAAAGTANAAAAATGGCCTTGCTGAGGAAATTAAATTTATGTTCAAGTGCTATTTCTTTACGGCACCGGGGAACAAGCATTTCAAACA
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| LTR10C | FoxB | 181 | 191 | - | 14.98 | GCAAATATTTA |
| LTR10C | THRB | 517 | 533 | - | 14.96 | TTTCCTCAGCAAGGCCA |
| LTR10C | RVE7 | 115 | 123 | + | 14.89 | TAAATATCT |
| LTR10C | NAC020 | 66 | 86 | - | 14.89 | TTTTGCTTCTAAGAGAAGTTA |
| LTR10C | LHY | 115 | 123 | - | 14.85 | AGATATTTA |
| LTR10C | THRB | 517 | 533 | + | 14.73 | TGGCCTTGCTGAGGAAA |
| LTR10C | CCA1 | 116 | 123 | + | 14.72 | AAATATCT |
| LTR10C | Spi1 | 234 | 246 | - | 14.71 | GAAGAGGAACAGG |
| LTR10C | SOX12 | 451 | 460 | - | 14.69 | ACCGAACAAA |
| LTR10C | RVE1 | 115 | 123 | + | 14.61 | TAAATATCT |
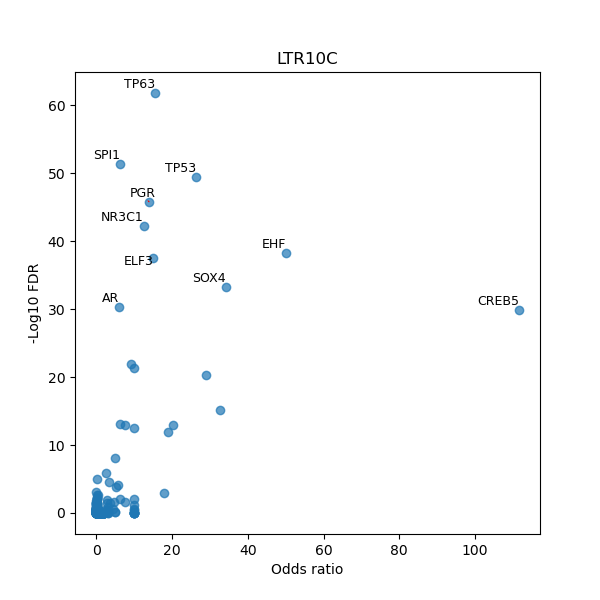
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.