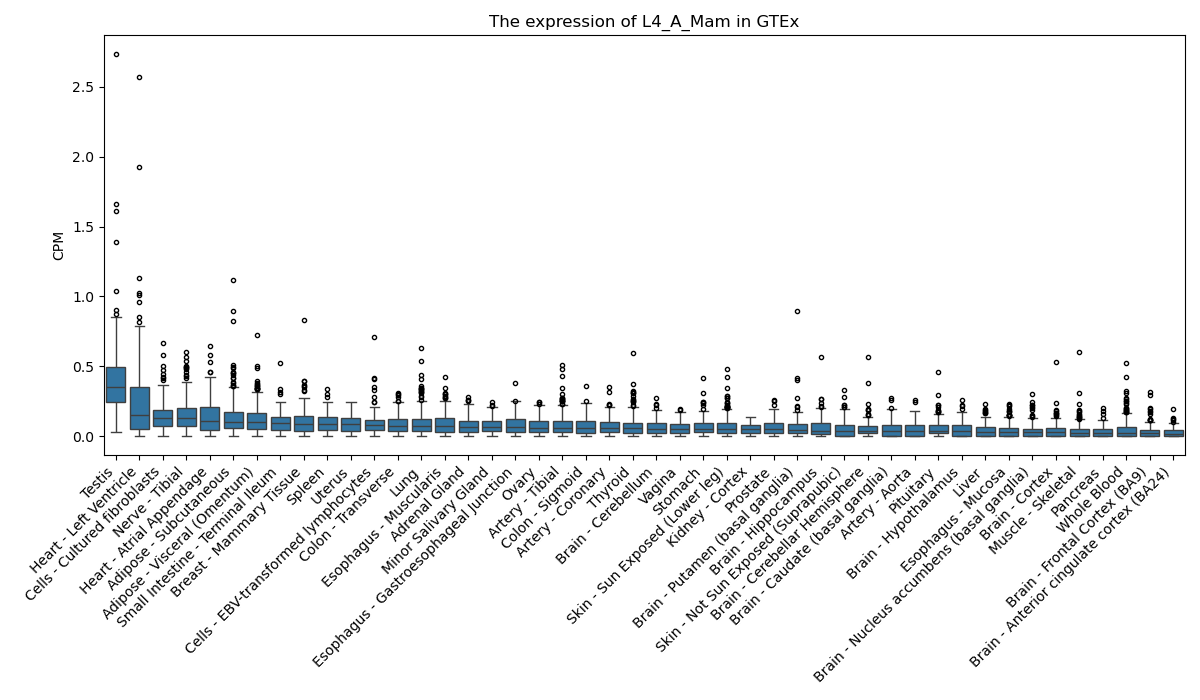
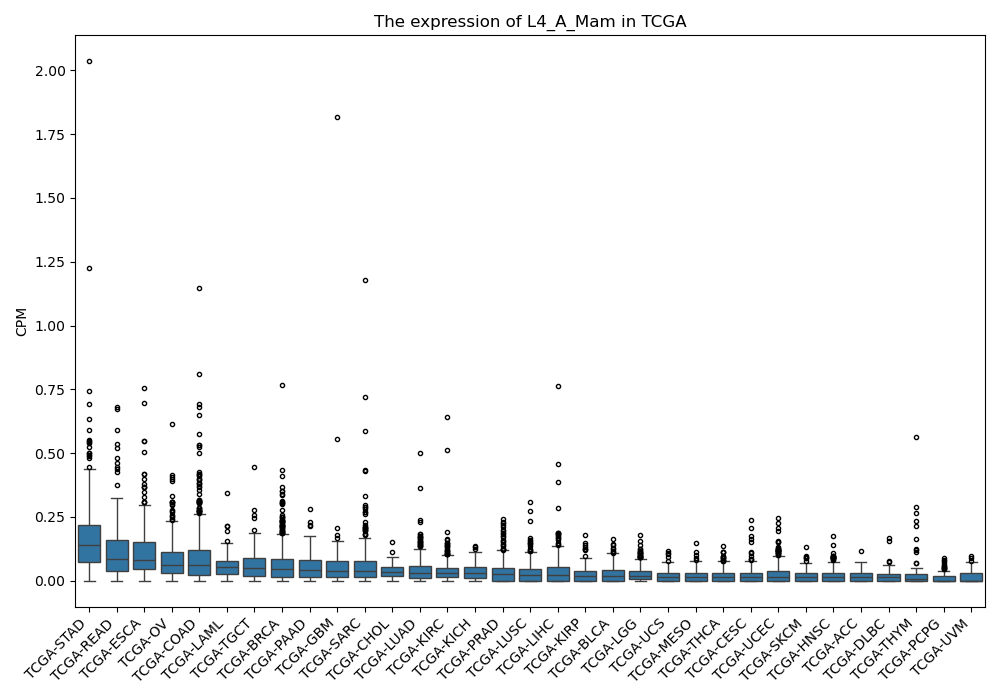
L4_A_Mam
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000364 |
|---|---|
| TE superfamily | RTE-X |
| TE class | LINE |
| Species | Eutheria |
| Length | 4990 |
| Kimura value | 35.46 |
| Tau index | 0.8282 |
| Description | RTE-X retrotransposon, L4_A_Mam subfamily |
| Comment | L4_A_Mam is one of three ~80% similar L4 subfamilies. ORF1 (3-1490) is not fully reconstructed, though ORF2 (~1491-4805) may be. These proteins similar to those of RTEX-1_ACar in lizards and L5 in ancient mammals. The substitution level found in the Laurasiatherian reconstruction is ~37%. |
| Sequence |
AGTGGTAAAAATGCAGACATCCTCACCCCAGACGGGAGNANTNNAGGAGCCCCTCTCCCNCAGCTGCCCCTGCAGCTGCCTCCCTGGCTCCCTCNAACGGATCTCCTAGCCCANTTAAGGATGGCTCTCCAAAAANCCTCACCCCGGGAGACTCGGGAGGCCCACGGGTTNATTCTCCCCAGGGCTATAATTCTCNCCGTGCAGATGGCACNTTTGATTTNCTGGAACGGTGTTNCTTCCTGACATCTCACACCATCCAGNCCTTATATGAGGAGCTGCGCGTTTTAAAATCCGTGGTCTCCAANATACATAACCATTGGATTTGGGGCAAACCGTGCCAANAGCATGCCACGAGNCAGGAGGGGATGGTGGACGGGAGGTCACACCCGGGGAAGGGCCCTCCATGCANNGCCCCTGGCCTGACCCTGGTGAGGAACCAGGTGGCGNTGAACCTCCCGATGGACNCTCATGGGAGGAGGCACGGTAGAGGNTCCGTAGCCAGGCTCCTGGNACCGCTTTTACACCAGCCTGCTTCTAATNCACTTATTTCCAGGTCGNCCCACCTCCCTTCCCGGCAGGGTCATAGGAGGANCCTCTTGGCGTGCGGCTCTCCCCAGCTCCCTTCCCTCATTATGANGGTCAAGGAGAANCTGGCTAGAGCGNGGNTTATCCCGAATAGGGTTTTTAAAACGGGGTCAGCTGCGCCCCTGCTGGGCGGTTCTCTTAATTTGTCAGGACCTNTACTGGGCAACCCAGGTCCGCCTAGAANACCAGAACCTAAGGAGAAGGGGAGGCCCGTTTCACCTCCAGACCATGGGCCTTGGTCCCCCCTGGGAAAGTTCTCGGGGCACAGGGAGGTGGCTGAGGCTGGGTTCTCTCGATCCTTTGTCCGTCTCCCTAAGGGAGAACAGGACGCTATTCTGGGGAGGTTAGAGTCCCTGAAGGATNANTTAATCAACCTTCAGGGCCTAGGCCATAATGANAATACCCCTGACTTCATGNAGTTTTCTCCCATCGGANCTCCTGTTGGGTCCCCTTGNAGGACNCCANCGAAGGGGAGCTCTCTCCCCNGAGNGGGCCCCGCCCCATTACTGCACAATCTGACTGATTGCACCTCCCACCCAGCCTCAGGAAGTGACATCTCCCCTAGCCCTTTCCTGACTGAGAGACTGGACCCTCAGGCCGCCCCCACTCATTGCCCGACTCCNCTCTCCCNAGTGATAGTCCTNGAGAATCCTCCTTGTTTGGCTGACTCACCAGTAAGCCCCACCCCAGTGCAGGTGACCTGGCCCAGAAAGCCTGCAGACTCCCCTATTTCCAGGAACTGGTTAGACATGCTCCACCTTGGNCCGGATGAGCTTGGCTTGCCAGATCCGGGGCGAGCCGCTCCGCCGGAAGCAGCGGCCTCGGCGGGTGCTAAGCCACACGGTAAGGCCAACGGCTGCATTCTTTTAGATCCGGTAGACTCCCTGGAGCGTGCCCCTCAATGACTAAGATTCCTCTCCTGGAACGTGGCCGNTTGGGGCCCTAAAGGTAAGGACCCCGATGTGTCGGCTTTTCTGATGGGTTTTAATATTATATGCCTGTAAGAGACCTGGATTTTAGAACATTCATCTCCCCATATTCANGGTTTTAGACCTTTTATCTCTCCGGCTGTCGGGGAAAAAAATTTTGGCAGAGGTAAGGGTGNATTGGCCACCCTTATCTCCGTCAACCTCAGGGGGACTGCTGTCGAGCTNCCTGGCTCATATGACAGGAATCTTTTCCTTCTGGTTCTGATTCGGTTCCCTGGCAAGCTGGATGTGATTTGTCTTAACACTTACATTCCCCCTGCCAAGGCGCACGCTGTCTGCTTGGACATATGGTCTCATTTTNATGATCTTTTAACTTTTATTTATTCTACGTACCCCATGGCGGAGTTAATCATCTCTGGGGACCTGAACGCCAGGATTGGCGGCGGGTCCGATGGGGCCCTACCTGGAGCCGCTGAGGAATGGGAGGATTGCTTCCCCGTGGGCCATTCTTTTAGAGACAGATGTATTAACCTAAATGGAAAATTTCTCACCAAACTTATTTATGAACAGAACCTGGTGGTGCTGAATGGTAGCACGTGGGATAAATCTGGGGGAAATTTTACCCGTATCTCTACTCTGGGGGCCAGCATCATAGATTATATCATAGTTAGCCCCTCTCTGCTTACTACCATCCTTAGAATGGATATTCTGGACCGGGTAGAAAGTGATCATTTCCCTCTCGTTTTAACCCTTGGCGTAGCTGCCCCGGAGCCCGTCTGCACTCATGATTGGACTGGNGAGATTAGGGGGCTGAGAAGAGTAAGATGGACCGAGGGGCTTTCAAACTCTATTGGTGACCTNTTGCTATCCGAGGACTTCTTAAAACTCTGCCTCAGGTGCCTCGAGGGAGAGGTTTCTCCTCTCATCTCTTATCAACGGATTGCTGATAACTTAAAACCGGTTTTATCCGCACTGTGTCCCGGCAAACGTGGGACTTACCCTCCTAGTGCCTGGTTTGACAAGGACTGTCAGGNAGCCAAAAAAATCTTGGCCAGGTTAGTGAGGCGACATGCGAAACGAAAATCGGAAGAGGCCACCCGGGATCTTATCAAATTTAAATCTTATTATAAACTTCTCATTGCCTCGAAAAAACTCAGGTATAATAAATCCNTGTGGGAGGACCTGAGTTTGGCCGTTAGATCCGGTAATGAGGGCAGATTCTGGGACATAGTTACCCGAGGTATGAATTTGGTGAGTGAGGTCGTGGAAGCTCAGATCCCGGTTGACACCTGGGAGGCTTACTTTTCCCAGCTTTACAAACCAAGGTCAACTGCCCCGAACTTTGTAGATCCCAGGCCGCTTTCTGACTGGGTCCCGGTAATACTCCCTCATATNGTACAGTTGATTAAAAAGCTCAAGCTAAATAAAGCACCCGGGGAAGATTTCCTGCCTCCAGAACTGTTCAAGGACCGCTCTGAATGGTGGGCTCCGATCCTTGCCAATTTGTTTACCTTTATCAACTCTACGGGTATGGTCCCGTCTGGTTGGACCCAGAGTGTAGTCTATCCCATTTTTTAAAAGGGCAATCCTTTACTTCCCCCAAATTATAGACCTATTAGCTTACTAGACATTCCATCTAAAATGTATGCCAGTTTCCTTCTTGATAAATTGCAAGTCTGGGTTTCTCAGGCCAATATTCTACACGAGGAGCAGGCGGGCTTTAGGCACGGCTATTCCACTATTGACCACTGTTATACTCTTTATCACCTTGTGGAGAAATCTGTCAGGAACAACATAAGATTGTTTGCGGCTTTTATTGACCTTTCCTCGGCCTTTGACTCTGTGGACAGGAATCGGTTATGGGCTAAGCTGCATGAGCTCAATATAGACCCCCGGCTATTGATGCTCATACAGAACCTGCACCTTAATACCACCGCGAGAGTCAGGGTCAGTAGGAACGGTCTCTTGACGGATCAGATAACAATTTCCAGTGGGGTAAAACAGGGATGTGTTCTGCCTACCCTCCTTTTTAACTTGTATCTTAATGACTTGATACCACTTTTGGATGAGCTGGATGCATGCCCTCCTGCCATAGAGAACAGAAAGATAAGCATTCTCCTATATGCAGATGACATGGTTTTATTGTCACGAACTAGGAGTGGCCTTAATAGACAACTGGCCCTGCTATCTAATTACTGCCAGAAAGAANGGCTTNAGATCAACTATTCTAAAACCAAAGTCATCATTTTTGGTAGACGTCCTCCAACATTTAACTGGCTTATAGCTAATAACCCTATACAGCAAGTCAACTCATTCAGTTACCTAGGNGTACATTTTGCAACTAGTTTATCTTGGAGGGTTCACCAGGAAGTTACATTGCTCAAAGTTAGACGTTCTATGGGTGCTTTACTGAGATTTTTTTATGGCCGAGGTGGGCGTTTGGTNACACCTGCTTTAAAAATTTTCCAGGCTAAAATTATTGCGGCCATGCTCTATGGCGTAGAACTTTGGGGCCTCGATCGCCCATTTGTCCGTGTGTTGGAGCAGACCCAGAACTGTTTTCTGAGGAAAATCTTGGCCCTGCCCGCGGGTACTCCCTCGGCCCACCTCCGTGCGGAGGTGGGGTGGCCTTCTATTCAGGCACGCGTCCTNGTTAGGCTCCTCAATTTTCATAAGAGGATGTCAACCCTACCCCCAGCTCGGCTGACTGTTAAAGCATATGGNTCTGCNCTTAACCGGCAACATAGAATAGCTGCACTCCAGGTNCTTGTCAGAGAGTATAACCTCGAACTCNCTGCCGCCCAATACTTATCAAAAGCTCGGTTGAGAGAAATAATATTTATGGAGGATTGCCTGAAGGATATGCAGTCCATCCATTCCTCTAGATACTCTAAANTCTATCCTTGGATTAAGCCAGACCANCAGAGAGCTGCGTACCTGGACCGCATTGTTTTGGCTCCCTGCAGAATTGCTTTTACTGAATTGCGCTTTGGCGTTATGCCATCGGCCTACATTGAGGGACGTTATAAGAAACAGCCCTATGAAACTCGTTACTGTATTTACTGTAAGGATGTTGTTGAGGACGTTGTTCATTACATCACACAGTGTCCTCTTTATGAGGACCCACGGGAGAAATTTCTCTCNGGTCTCAGTACCAGGAANAGCTGTGCTTCTCCTGAGCAACTGGTTTGTTTTTATCTTATGGACACTGTGAATCGTGTAACTGATCATGTTTCCCTCTTTGCNTTGGCTGCTAGGAAGCTCAGAGCCAAATTTGTGGCCCACCTCTAGCAANTGTACAGGACCTTATGGCATGCATTTTGTGTACTAACCTCACTCCTGGCTTCATCTTATTTTTCTACCCTTATTTTCCTTTTTCCTGATTTTTTAAATTTATATTTTTTAATTTTATTGTATGTATTTTTGTAAGCCGCCTTAAATCCTTTTTGGAACAAGGCAGGATATAAATAAATAAA
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| L4_A_Mam | ZNF684 | 786 | 799 | - | 16.54 | ACGGGCCTCCCCTT |
| L4_A_Mam | Sox14 | 881 | 891 | + | 16.51 | ATCCTTTGTCC |
| L4_A_Mam | TCX6 | 2627 | 2641 | + | 16.49 | ATTTAAATCTTATTA |
| L4_A_Mam | Stat92E | 1315 | 1326 | - | 16.47 | AGTTCCTGGAAA |
| L4_A_Mam | CDX1 | 3933 | 3942 | - | 16.42 | GGCCATAAAA |
| L4_A_Mam | ERF::FIGLA | 431 | 443 | + | 16.39 | GAGGAACCAGGTG |
| L4_A_Mam | KLF15 | 1079 | 1086 | + | 16.27 | CCCCGCCC |
| L4_A_Mam | ERF082 | 1952 | 1960 | - | 16.26 | CCGCCGCCA |
| L4_A_Mam | Ikzf3 | 1129 | 1137 | + | 16.18 | CAGGAAGTG |
| L4_A_Mam | ERF4 | 1952 | 1960 | - | 16.16 | CCGCCGCCA |
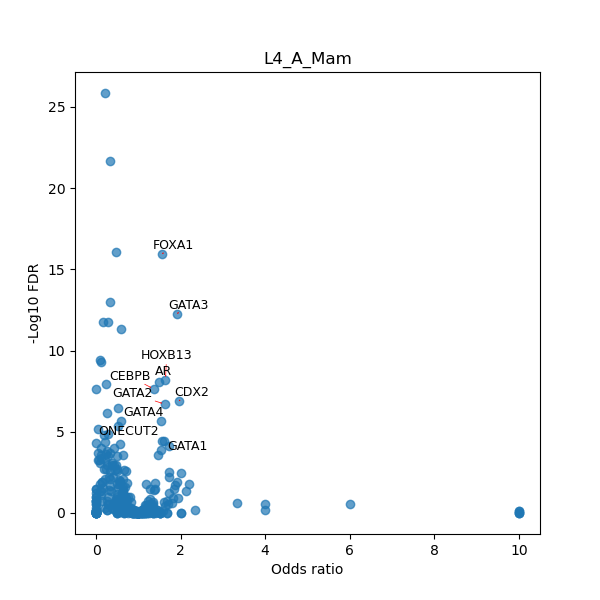
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.