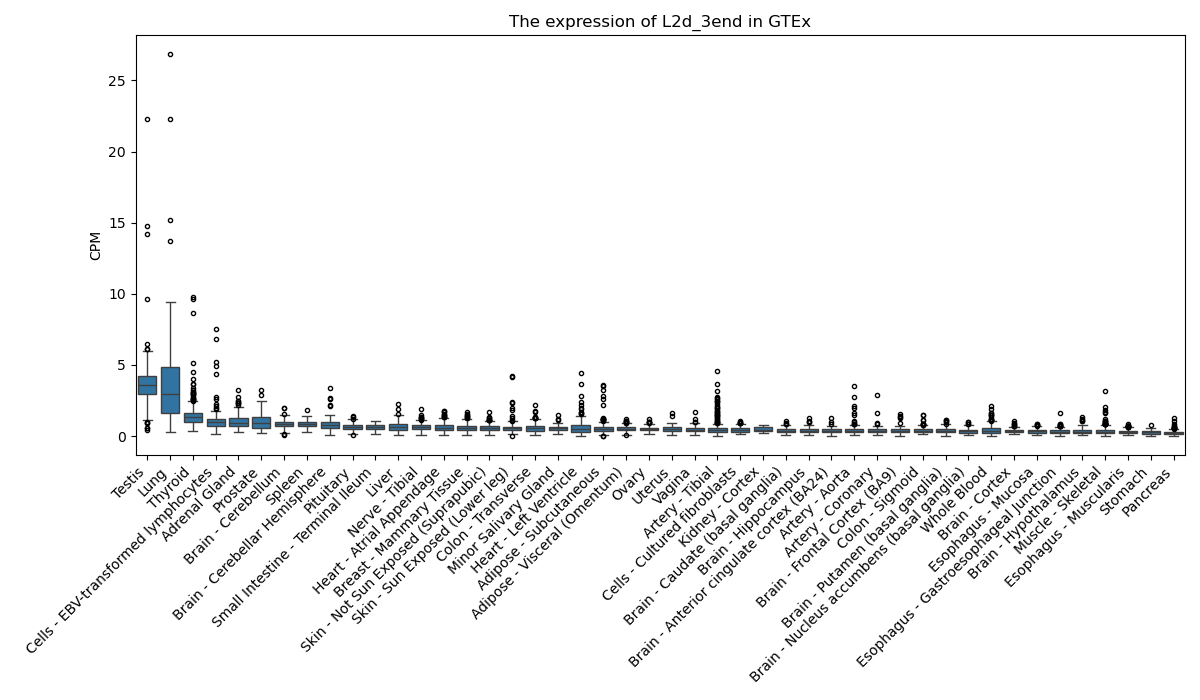
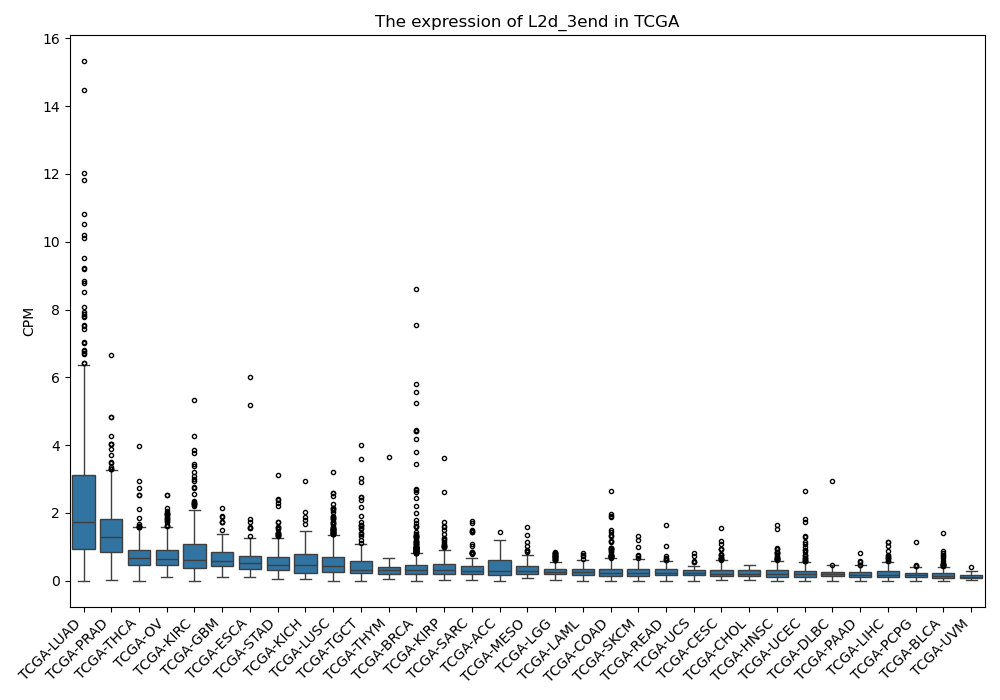
L2d_3end
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0001215 |
|---|---|
| TE superfamily | L2 |
| TE class | LINE |
| Species | Eutheria |
| Length | 516 |
| Kimura value | 36.41 |
| Tau index | 0.8395 |
| Description | 3' end of L2 (LINE2) retrotransposon, L2d_3end subfamily |
| Comment | Copies at orthologous sites in all eutheria. |
| Sequence |
CTCCACGATCTGGCCCCACCCTACCTNTCCAGCCTTATCTCCCACTACTCCCCAGCACGAACCCTCCGCTCCAGTCAGGCCGGTCTCTTCACTGTCCCCCGAACACGCCTTGCGCATTCCCGCCTCTGCGCCTTTGCTCACGCCGTTCCCCCCGCCTGGAATGCCCTTCCCCTTCCTCTCTCTCTATCCAAATCCTACCCATCCTTCAAGGCCCAGCTCAAGTCCCACCTCCTCCACGAAGCCTTCCCTGACCACTCCAGCCCACACTGATCTCTCCCTTCTCTGAACTCCTATAGCACTTATAGTCTGTACCACGCAATTTAGCACTTAATTATATACTGTCTTGTATTGTTCTCTAATTGTTTCATGTGTGTTAGTCTTGTCTCCCCAACTAGATTGTAAGCTCCTTGAGGGCAGGGACCATGTCTTATACTNTTTTNTCGTTCCCCACAGCGCCTAGCACAGTGCTGAGCACATAGTAGGCGCTCAATAAATACTTGTTGATTGATTGATTGA
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| L2d_3end | KLF1 | 14 | 21 | - | 14.64 | GGGTGGGG |
| L2d_3end | MAC1 | 133 | 140 | + | 14.47 | TTTGCTCA |
| L2d_3end | ZKSCAN1 | 476 | 484 | + | 14.33 | ATAGTAGGC |
| L2d_3end | Wt1 | 228 | 237 | + | 14.31 | CCTCCTCCAC |
| L2d_3end | TCP5 | 417 | 424 | + | 14.30 | GGGACCAT |
| L2d_3end | Glyma19g26560.1 | 12 | 19 | + | 14.23 | GGCCCCAC |
| L2d_3end | GLYMA19G26560 | 12 | 19 | + | 14.23 | GGCCCCAC |
| L2d_3end | GATA4 | 33 | 40 | + | 14.20 | CCTTATCT |
| L2d_3end | DREB2G | 225 | 238 | - | 14.20 | CGTGGAGGAGGTGG |
| L2d_3end | ARF36 | 377 | 388 | - | 14.18 | GGGAGACAAGAC |
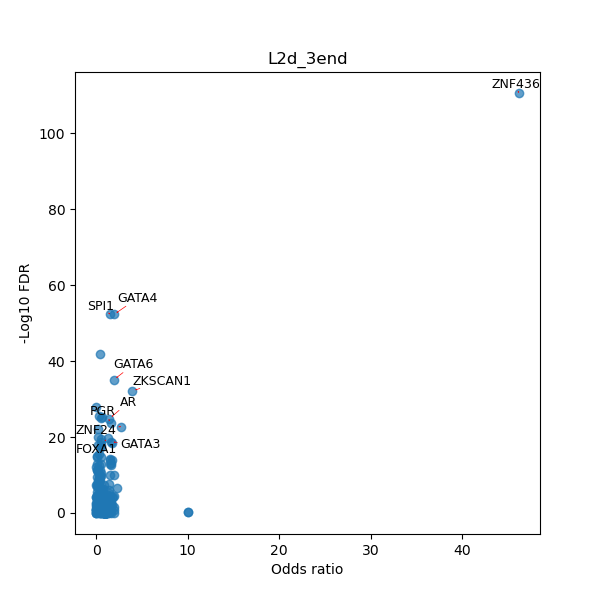
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.