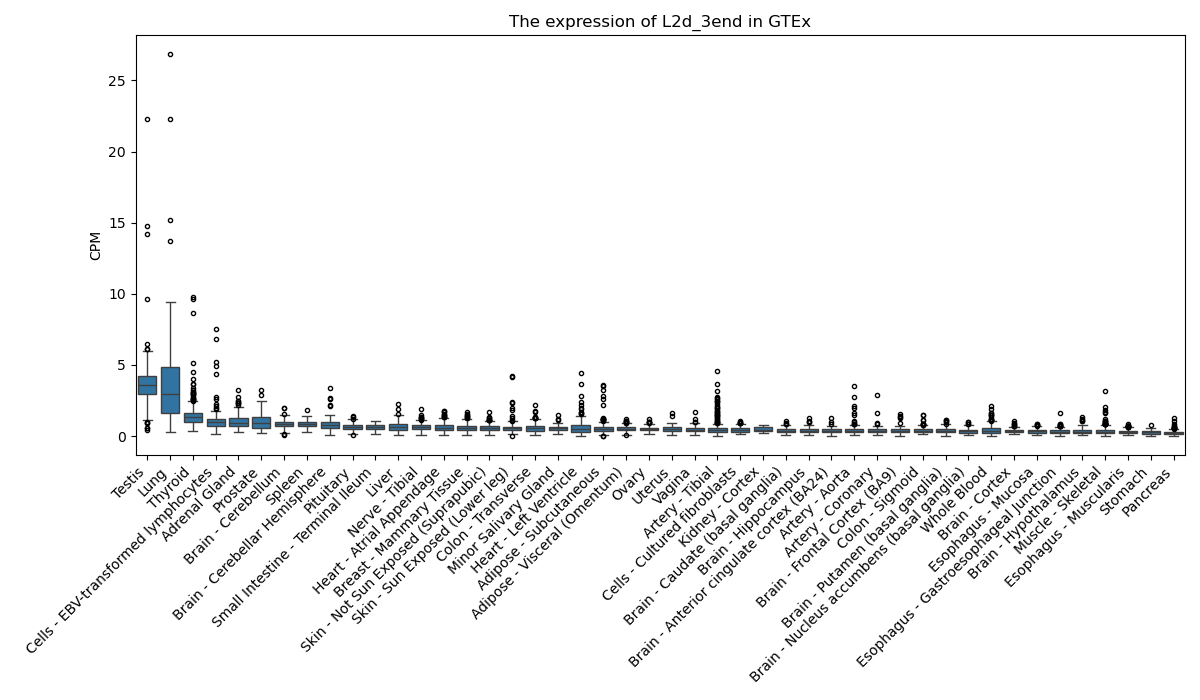
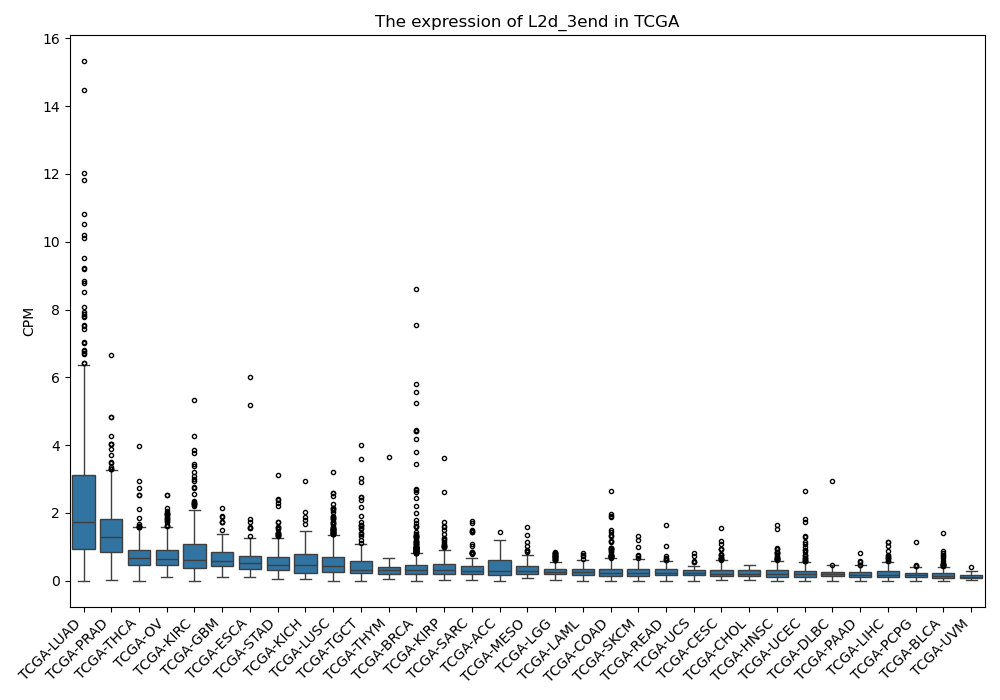
L2d_3end
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0001215 |
|---|---|
| TE superfamily | L2 |
| TE class | LINE |
| Species | Eutheria |
| Length | 516 |
| Kimura value | 36.41 |
| Tau index | 0.8395 |
| Description | 3' end of L2 (LINE2) retrotransposon, L2d_3end subfamily |
| Comment | Copies at orthologous sites in all eutheria. |
| Sequence |
CTCCACGATCTGGCCCCACCCTACCTNTCCAGCCTTATCTCCCACTACTCCCCAGCACGAACCCTCCGCTCCAGTCAGGCCGGTCTCTTCACTGTCCCCCGAACACGCCTTGCGCATTCCCGCCTCTGCGCCTTTGCTCACGCCGTTCCCCCCGCCTGGAATGCCCTTCCCCTTCCTCTCTCTCTATCCAAATCCTACCCATCCTTCAAGGCCCAGCTCAAGTCCCACCTCCTCCACGAAGCCTTCCCTGACCACTCCAGCCCACACTGATCTCTCCCTTCTCTGAACTCCTATAGCACTTATAGTCTGTACCACGCAATTTAGCACTTAATTATATACTGTCTTGTATTGTTCTCTAATTGTTTCATGTGTGTTAGTCTTGTCTCCCCAACTAGATTGTAAGCTCCTTGAGGGCAGGGACCATGTCTTATACTNTTTTNTCGTTCCCCACAGCGCCTAGCACAGTGCTGAGCACATAGTAGGCGCTCAATAAATACTTGTTGATTGATTGATTGA
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| L2d_3end | RAMOSA1 | 175 | 188 | - | 19.37 | GATAGAGAGAGAGG |
| L2d_3end | BPC1 | 165 | 188 | - | 18.27 | GATAGAGAGAGAGGAAGGGGAAGG |
| L2d_3end | BPC6 | 168 | 188 | + | 16.50 | TCCCCTTCCTCTCTCTCTATC |
| L2d_3end | PBX1 | 501 | 509 | - | 15.97 | ATCAATCAA |
| L2d_3end | PBX1 | 505 | 513 | - | 15.97 | ATCAATCAA |
| L2d_3end | KLF5 | 13 | 22 | + | 15.96 | GCCCCACCCT |
| L2d_3end | RAMOSA1 | 173 | 186 | - | 15.77 | TAGAGAGAGAGGAA |
| L2d_3end | ZHD9 | 320 | 334 | + | 15.64 | TTTAGCACTTAATTA |
| L2d_3end | Spi1 | 169 | 181 | - | 15.50 | AGAGAGGAAGGGG |
| L2d_3end | KLF4 | 14 | 21 | + | 15.48 | CCCCACCC |
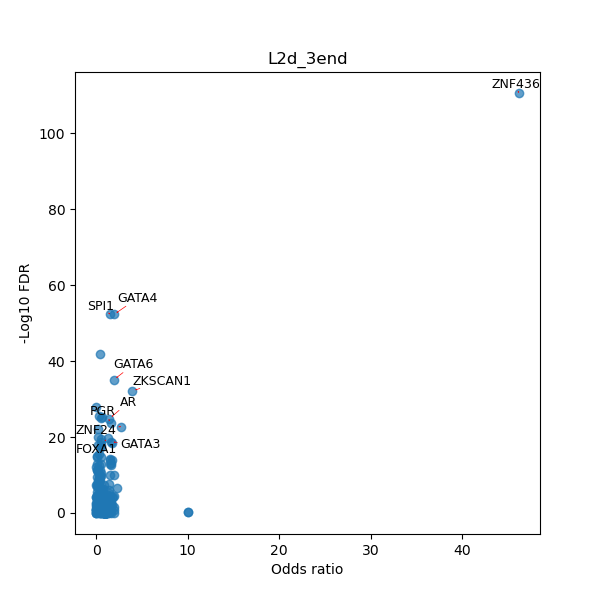
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.