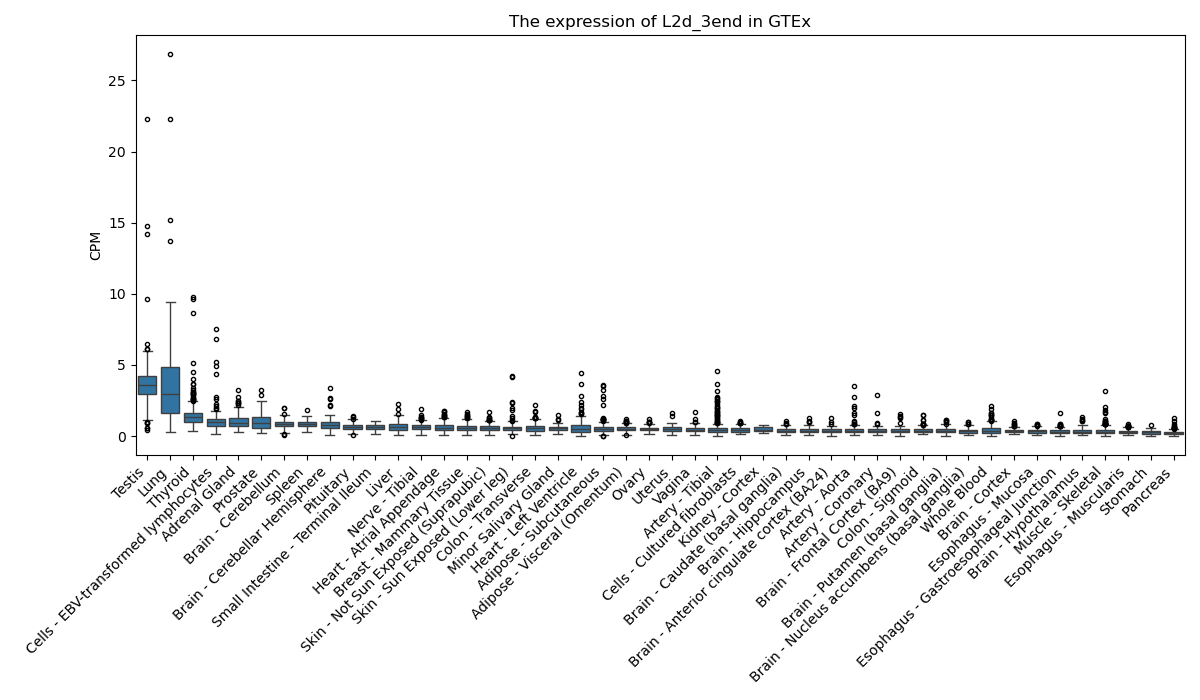
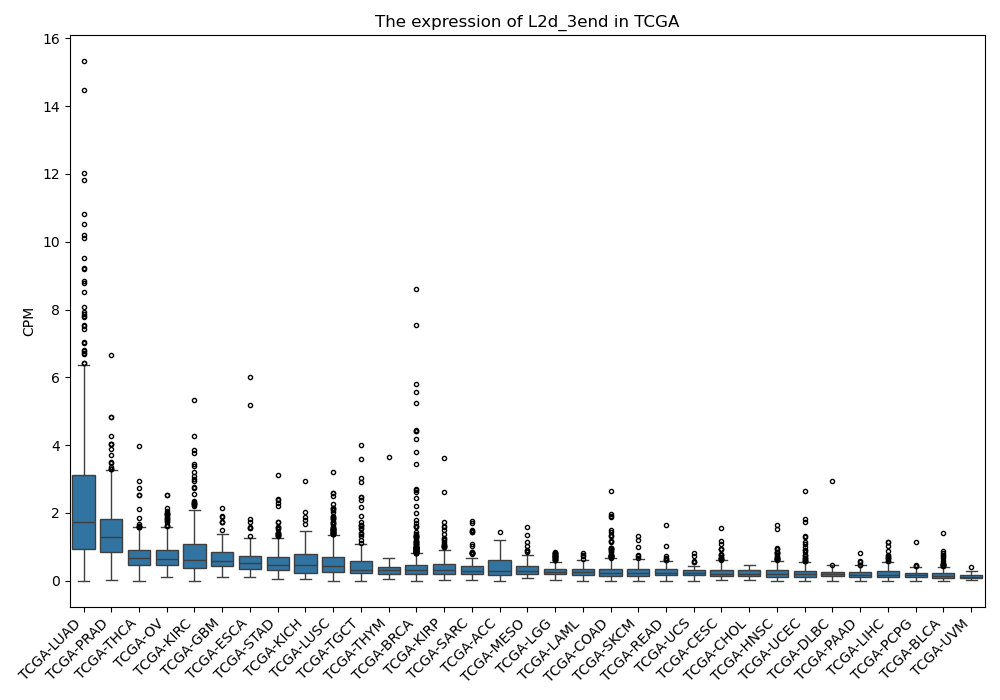
L2d_3end
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0001215 |
|---|---|
| TE superfamily | L2 |
| TE class | LINE |
| Species | Eutheria |
| Length | 516 |
| Kimura value | 36.41 |
| Tau index | 0.8395 |
| Description | 3' end of L2 (LINE2) retrotransposon, L2d_3end subfamily |
| Comment | Copies at orthologous sites in all eutheria. |
| Sequence |
CTCCACGATCTGGCCCCACCCTACCTNTCCAGCCTTATCTCCCACTACTCCCCAGCACGAACCCTCCGCTCCAGTCAGGCCGGTCTCTTCACTGTCCCCCGAACACGCCTTGCGCATTCCCGCCTCTGCGCCTTTGCTCACGCCGTTCCCCCCGCCTGGAATGCCCTTCCCCTTCCTCTCTCTCTATCCAAATCCTACCCATCCTTCAAGGCCCAGCTCAAGTCCCACCTCCTCCACGAAGCCTTCCCTGACCACTCCAGCCCACACTGATCTCTCCCTTCTCTGAACTCCTATAGCACTTATAGTCTGTACCACGCAATTTAGCACTTAATTATATACTGTCTTGTATTGTTCTCTAATTGTTTCATGTGTGTTAGTCTTGTCTCCCCAACTAGATTGTAAGCTCCTTGAGGGCAGGGACCATGTCTTATACTNTTTTNTCGTTCCCCACAGCGCCTAGCACAGTGCTGAGCACATAGTAGGCGCTCAATAAATACTTGTTGATTGATTGATTGA
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| L2d_3end | ARALYDRAFT_495258 | 12 | 19 | + | 15.43 | GGCCCCAC |
| L2d_3end | ARALYDRAFT_484486 | 12 | 19 | + | 15.43 | GGCCCCAC |
| L2d_3end | ZNF530 | 162 | 175 | - | 15.40 | GAAGGGGAAGGGCA |
| L2d_3end | E2F6 | 117 | 124 | - | 15.29 | GGCGGGAA |
| L2d_3end | BPC6 | 170 | 190 | + | 15.22 | CCCTTCCTCTCTCTCTATCCA |
| L2d_3end | Plagl1 | 11 | 18 | - | 15.15 | TGGGGCCA |
| L2d_3end | PATZ1 | 149 | 159 | - | 14.89 | CCAGGCGGGGG |
| L2d_3end | Trl | 176 | 184 | - | 14.81 | GAGAGAGAG |
| L2d_3end | TFDP1 | 117 | 124 | - | 14.70 | GGCGGGAA |
| L2d_3end | ARALYDRAFT_493022 | 12 | 19 | + | 14.64 | GGCCCCAC |
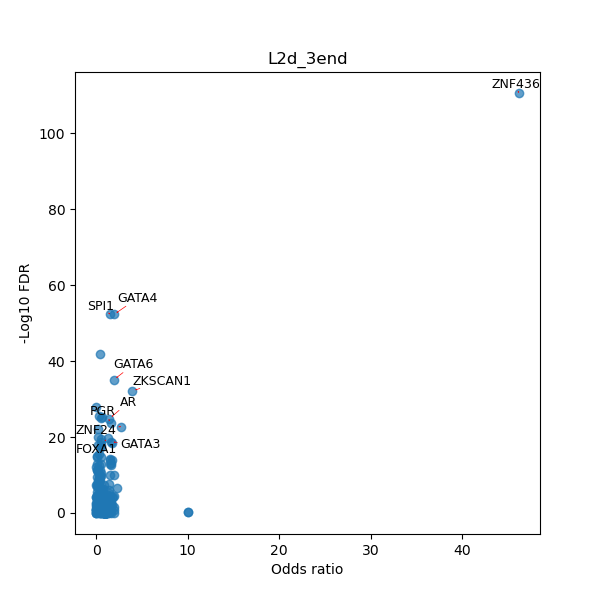
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.