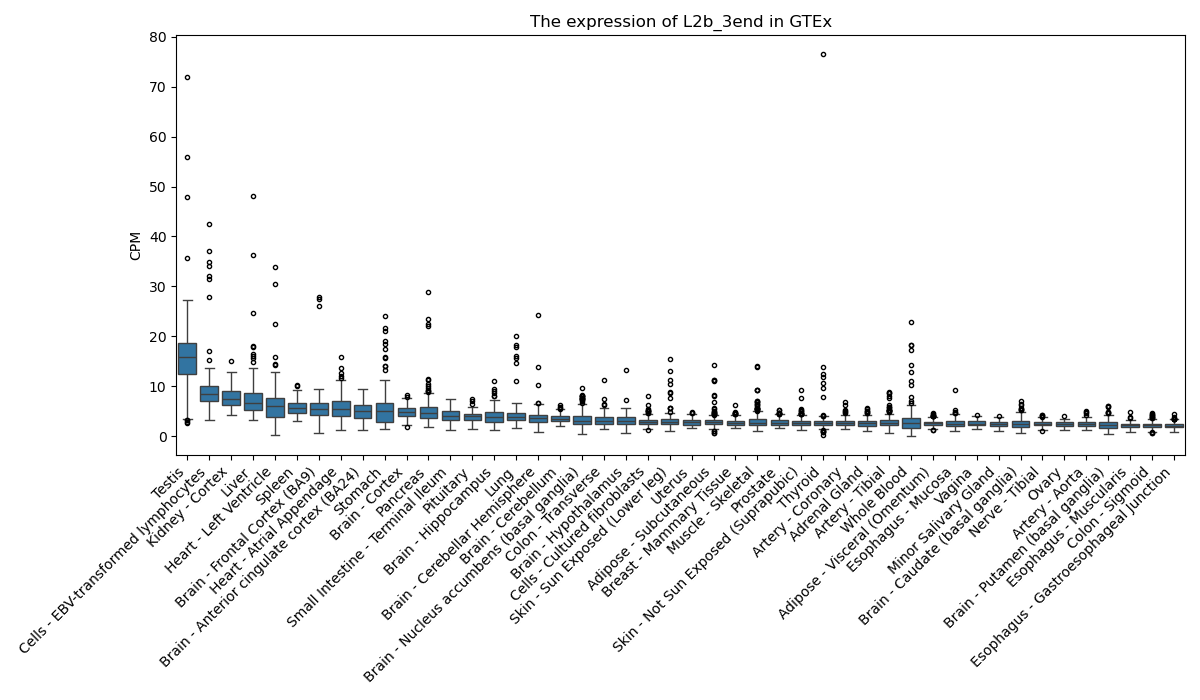
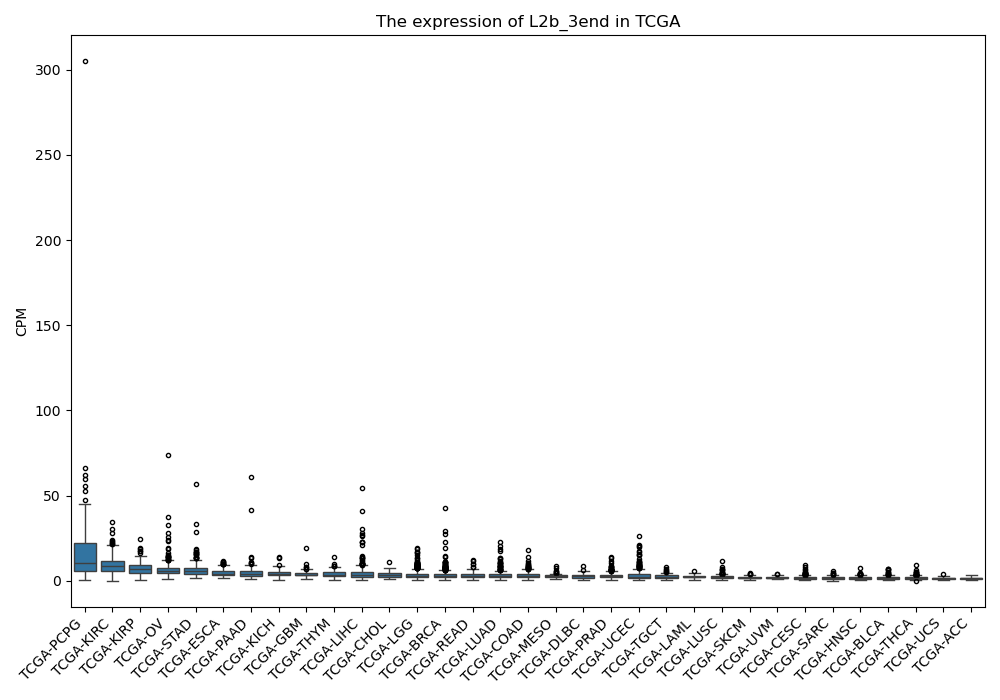
L2b_3end
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000360 |
|---|---|
| TE superfamily | L2 |
| TE class | LINE |
| Species | Theria_mammals |
| Length | 466 |
| Kimura value | 32.92 |
| Tau index | 0.7773 |
| Description | 3' end of L2 (LINE2) retrotransposon, L2b_3end subfamily |
| Comment | L2 3' region. Starts at 2910 of L2 consensus with ~170 bp overlap. MIR and MIRb share 3' terminal end homology over ~50bp. |
| Sequence |
TCGCACGATCTGGCCCCGCTACCTCTCCGGCCTCATCTCCTACCACTCTCCCCTTGCTCACTCTGCTCCAGCCACACTGGCCTCCTTGCTGTTCCTCGAACACGCCANGCTCNCTCCCGCCTCAGGGCCTTTGCACNTGCTGTTCCCTCTGCCTGGAACGCCCTTCCCCACCTCTTCGCCTGGCCAACTCCTACTCATCCTTCAGGTCTCAGCTCAAATGTCACCTCCTCCGGGAAGCCCTCCCTGACCCCCCAGGCCGGGTCAGGCGCCCTCCTCTGGGCCCCCCCAGTCCTACCCTGCCACTCTGGGTTATNATTGTCTGNTTACATGTCTGTCTCCCCCACTAGACTGTGAGCTCCGTGAGGGCAGGGACCGNGTCTGTCTTGTTCACCGCTGTATCCCCAGCGCCTAGCACAGNGCCTGGCACACAGTAGGCGCTCAGTAAATATTTGTTGAATGAATGAAT
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| L2b_3end | TB1 | 278 | 286 | + | 17.38 | GGGCCCCCC |
| L2b_3end | KLF17 | 42 | 55 | + | 17.27 | ACCACTCTCCCCTT |
| L2b_3end | TB1 | 279 | 287 | + | 16.64 | GGCCCCCCC |
| L2b_3end | ZNF189 | 138 | 146 | + | 16.13 | TGCTGTTCC |
| L2b_3end | ZNF189 | 87 | 95 | + | 16.13 | TGCTGTTCC |
| L2b_3end | TCP1 | 276 | 286 | + | 15.85 | CTGGGCCCCCC |
| L2b_3end | ZNF213 | 250 | 261 | + | 15.80 | CCCCAGGCCGGG |
| L2b_3end | FoxB | 442 | 452 | + | 15.70 | GTAAATATTTG |
| L2b_3end | Wt1 | 335 | 344 | + | 15.69 | TCTCCCCCAC |
| L2b_3end | PLAGL2 | 278 | 285 | + | 15.67 | GGGCCCCC |
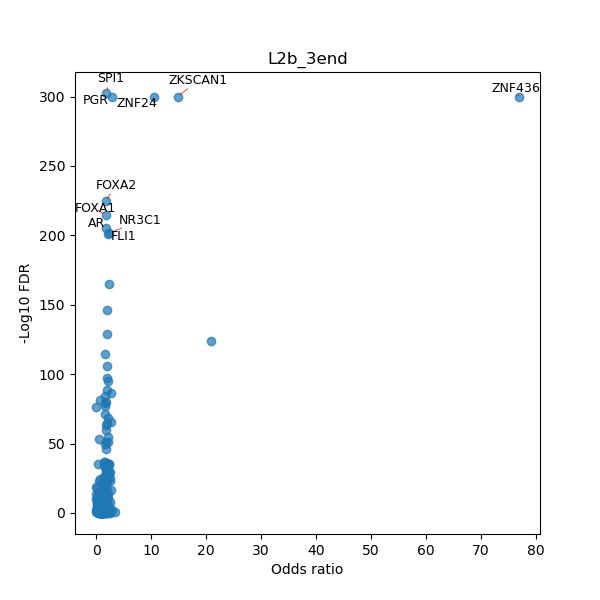
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.