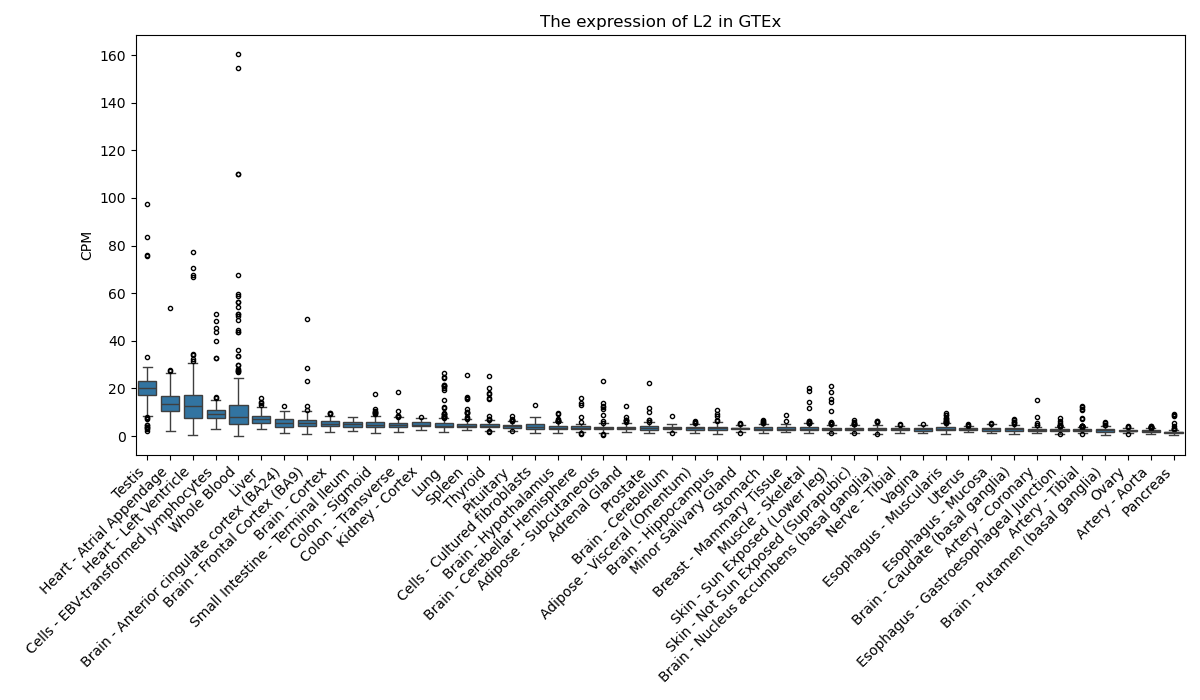
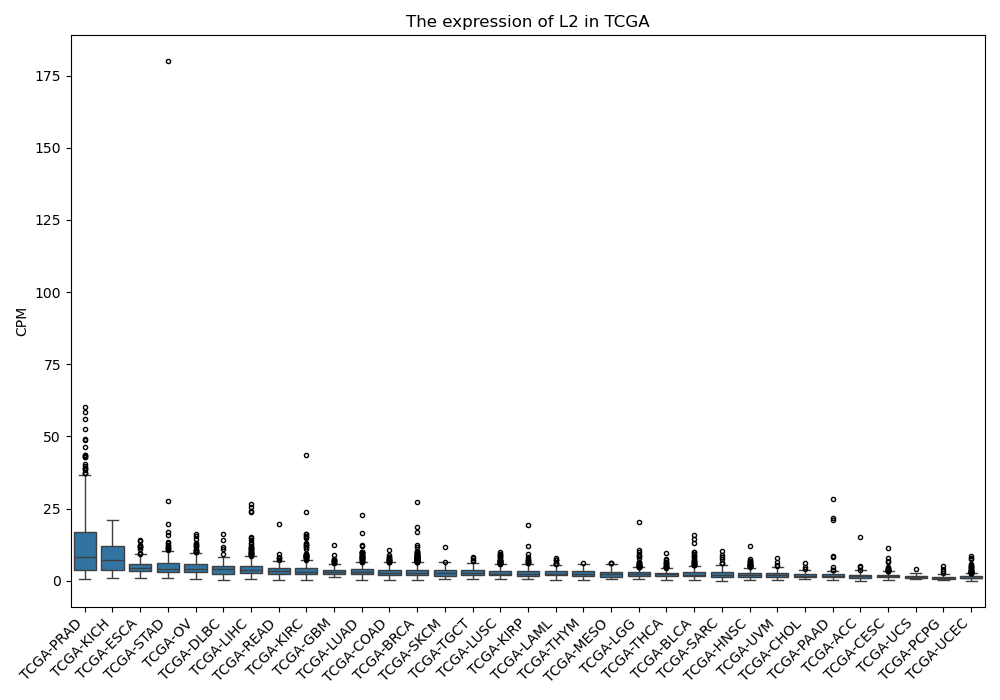
L2
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000358 |
|---|---|
| TE superfamily | L2 |
| TE class | LINE |
| Species | Mammalia |
| Length | 3082 |
| Kimura value | 37.63 |
| Tau index | 0.7930 |
| Description | L2 (LINE2) retrotransposon |
| Comment | L2 (LINE2) spread before the mammalian radiation, and copies are only 65-75% similar to the consensus. The 5' end is probably incomplete. The 3' end is also incomplete, but this terminus is extended by three model variants: L2a_3end, L2b_3end, and L2c_3end. Note that, whereas L1 is A- and purine-rich in the coding strand, L2 is C- and pyrimidine-rich. |
| Sequence |
ACCTCCNAGCCTCCATCGGCAGGGGTCAAGAGGGAGCCCCATGCGCTCNGAGTAAGACCGCCCTCCCCNGGCCTCCACGCCNCCATCGGCAGGAGTNAAGAGAGGGCCCCAGAACTCTGCGTGCTCCTGGCAAGACTCCNACTCCCCACTTCCCCCTCAACACTCACAGCCTCGCCACCACTATCTGCCTTGAGACCATCAGGACTAATCTCATCATGCCCTTTATCCCTGTTTACGTCAGTCACCACAAGAAGCCCAAGACTCCCAGGTCATCCCTCATCCCCCGGACTCTNATCCCCATACGGCTCTGTACATCTCCTNTTCTGACCCCATCTCCCACCCCTCCCCAACTCCTGAAACCCTTCCACTGTGCCCTCTGGAACTCACGGTCCGTCATCAGCAAAATCCCCCGTATCCTCAACCTCTTCTCTGAACGTTCCCTTCACCTTCTTGCTCTAACNGAAACCTGGCTCTCCCCTGAGGACACTGCTTCCCCTGCAGCCCTCTCAAGTGGTGGCTGTTTTCTCTCCCACANCCCTCGTACCACTGGGCCTGGAGGTGGGGTAGGTGTCCTCCTTGCTCCTCATTGCCGCTTCCAGACCATTCTCCCTCCCTCCTCCCTAAAACACCCCAGCTTTGAATCTCATGTCATCAGACTATACCACCCGCTACCCCTCCTTGTTGCAGTCATCTACCGACCTCCGGGTCACTCCCCCTCATTCCTTGAAGATTTTAGCTCCTGGCTCACTGTCACTCTCTCCAACACTACTCCTGTCATAATTCTTGGTGATTTCAATATCCACGTAGATGATCCTTCCAATACCCTGGCCTCTCAGTTCCTTGACCTCCTCTCCTCCAATGATCTTGTCCTCCACCCTACCTCAGCCACTCACTCCCATGGTCATACCCTAGACCTTGTCATTACCAATAACTGCAACCCCTCCATAATCTCAATTTCAAGCATCCCACTCTCTGACCACCACCTCCTATCTTTCCAGCTCACTCCCTCTAGTACCCCGACTCCAACAATTCTTCGACCCCACCGGGACCTCCAATCCATTGATCCTACCACCTTTTCACTGTCCCTCACCCCCCTCATGTCCTCACTTCCCTCCTTACCCAGCTTAGATTCCATGGTCNATCATTATAATCACTCCCTTGCATATACCCTCAACTCCCTTGCCCCTCTCTCGCTTCGTCGTACTCGCCTGGCAAAACCNCAACCCTGGTTAAATCCAACTCTCCGCCTACTCCGCGCCTGCACCCGNGCAGCTGAACGTGGCTGGAGAAAAACACACAACCATGCTGACTGGTCTCACTTTAAATTCATGACCACNAACCTCAAGTGGGCCCTTAATGCTGCCCGGCAATCNTACTACATTTCCCTAGTCCATTCACTCTCCCACTCTCCTAGACGACTATTTCACACCTTCTCCTCTCTCCTCAAACCTCCAACACCTCCTCCCCTATCCTCACTCTCAGCTGATGACCTTGCTTCCTATTTCACTGAGAAAATAGAAGCAATCAGAAGAGAACTTCCACANNCTCCCACCACCACATCTACCCACCTACCTGCATCTGTGCCCACATACTCTGCCTTCCCTCCTGTTACTACGGATGAACTGTCCGTGCTCCTATCTAAGGCCAACCCCTCCACTTGTGCACTAGATCCCATCCCCTCTCGCCTACTCAAGGACATCGCTCCAGCAATTCTCCCCTCTCTCTCCTGCATCATCAATTTTTCCCTCTCTACTGGATCATTCCCATCAGCATACAAACATGCTGTNATTTCTCCCATCTTTAAAAAACAAAAATTCTCCCTTGACCCCACNTCCCCCTCCAGCTACCGCCCCATTTCTCTGCTCCCCTTTACAGCAAAACTCCTCGAAAGAGTTGTCTATACTCGCTGTCTCCAATTCCTCTCCTCCCATTCTCTCTTGAACCCACTCCAATCAGGCTTTCGTCCCCACCACTCCACCGAAACTGCTCTTGTCAAGGTCACCAATGACCTCCACGTTGCTAAATCCAATGGTCAATTCTCAGTCCTCATCTTACTTGACCTNTCAGCAGCATTTGACACAGTTGATCACTCCCTCCTTCTTGAAACACTTTCTTCACTTGGCTTCCAGGACACCACACTCTCCTGGTTTTCCTCCTACCTCACTGGCCGCTCCTTCTCAGTCTCCTTTGCTGGTTCCTCCTCATCTCCCCGACCTCTNAACGTTGGAGTGCCCCAGGGCTCAGTCCTTGGACCTCTTCTCTTCTCTATCTACACTCACTCCCTNGGTGATCTCATCCAGTCTCATGGCTTTAAATACCATCTATATGCTGATGACTCCCAAATTTATATCTCCAGCCCGGACCTCTCCCCTGAACTCCAGACTCGTATATCCAACTGCCTACTCGACATCTCCACTTGGATGTCTAATAGGCATCTCAAACTTAACATGTCCAAAACTGAACTCCTGATCTTCCCCCTCAAACCTGCTCCTCCCGCAGTCTTCCCCATCTCAGTNAATGGCAACTCCATCCTTCCAGTTGCTCAGGCCAAAAACCTTGGAGTCATCCTTGACTCCTCTCTTTCTCTCACACCCCACATCCAATCCATCAGCAAATCCTGTCGGCTCTACCTTCAAAATATATCCAGAATCCGACCACTTCTCACCACCTCCACTGCCACCACCCTGGTCCAAGCCACCATCATCTCTCGCCTGGATTACTGCAATAGCCTCCTAACTGGTCTCCCTGCTTCCACCCTTGCCCCCCTNCAGTCTATTCTCAACACAGCAGCCAGAGTGATCCTTTTAAAACGTAAGTCAGATCATGTCACTCCTCTGCTCAAAACCCTCCAATGGCTTCCCATCTCACTCAGAGTAAAAGCCAAAGTCCTTACAATGGCCTACAAGGCCCTACATGATCTGGTCCCCCGTTACCTCTCTGACCTCATCTCCTACCACTCTCCCCCTCGCTCACTCCGCTCCAGCCACACTGGCCTCCTTGCTGTTCCTCGAACACGCCAGGCACGCTCCCGCCTCAGGGCCTTTGCACTTGCTGTTCCCTCTGCCTGGAACGCTCTTCCCCC
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| L2 | EOMES | 1423 | 1431 | - | 15.76 | AGGTGTGAA |
| L2 | OsI_08196 | 1346 | 1353 | - | 15.69 | GGGCCCAC |
| L2 | eor-1 | 2575 | 2587 | - | 15.68 | AGAGAAAGAGAGG |
| L2 | NR2F1 | 21 | 32 | + | 15.67 | AGGGGTCAAGAG |
| L2 | MYB74 | 1564 | 1574 | + | 15.67 | CCCACCTACCT |
| L2 | Tbx6 | 1423 | 1431 | - | 15.67 | AGGTGTGAA |
| L2 | Ebf4 | 474 | 484 | - | 15.67 | TCCTCAGGGGA |
| L2 | ARALYDRAFT_493022 | 1346 | 1353 | - | 15.63 | GGGCCCAC |
| L2 | ZNF148 | 340 | 349 | + | 15.61 | CCCCTCCCCA |
| L2 | MYB51 | 1564 | 1574 | + | 15.59 | CCCACCTACCT |
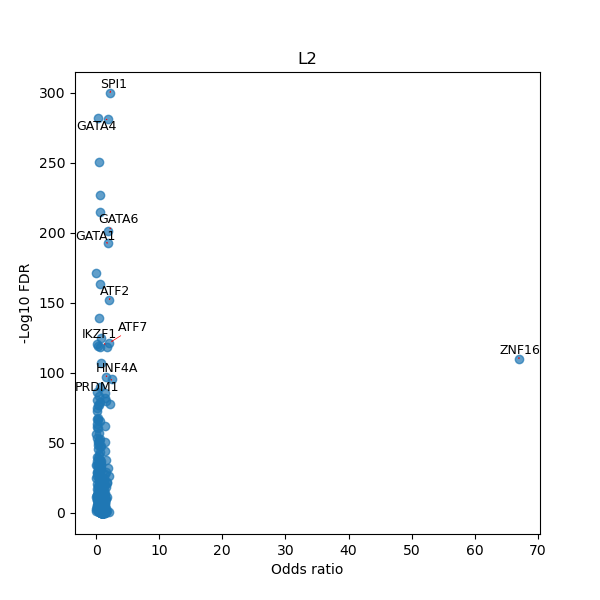
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.