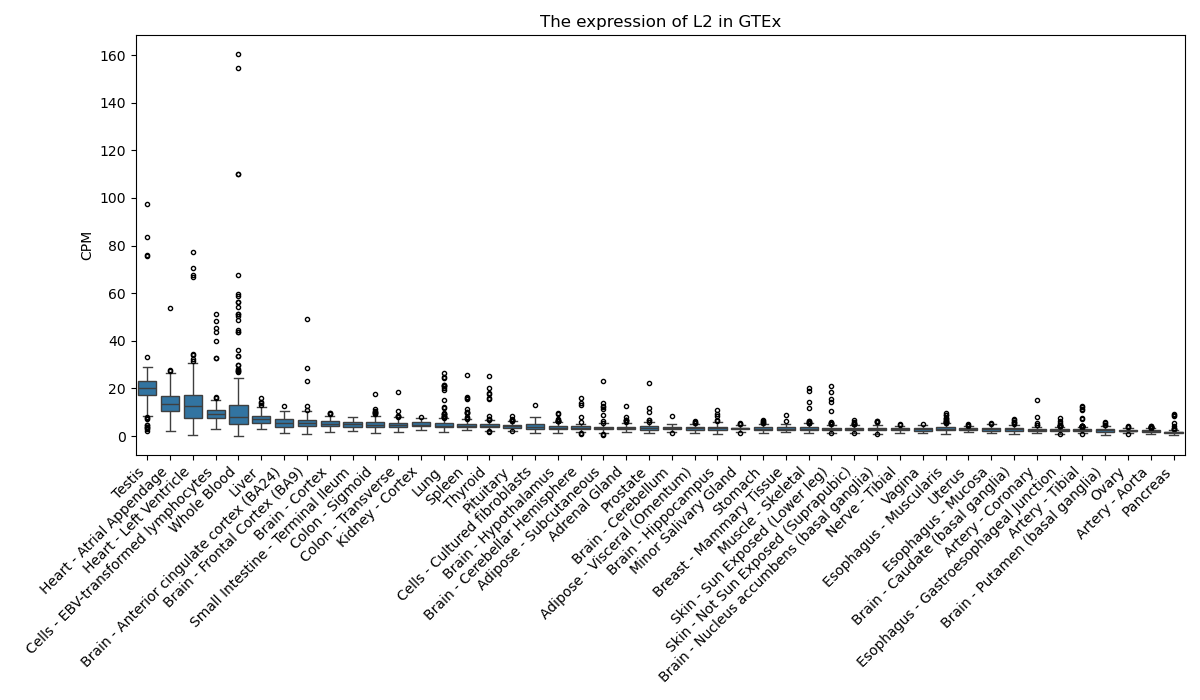
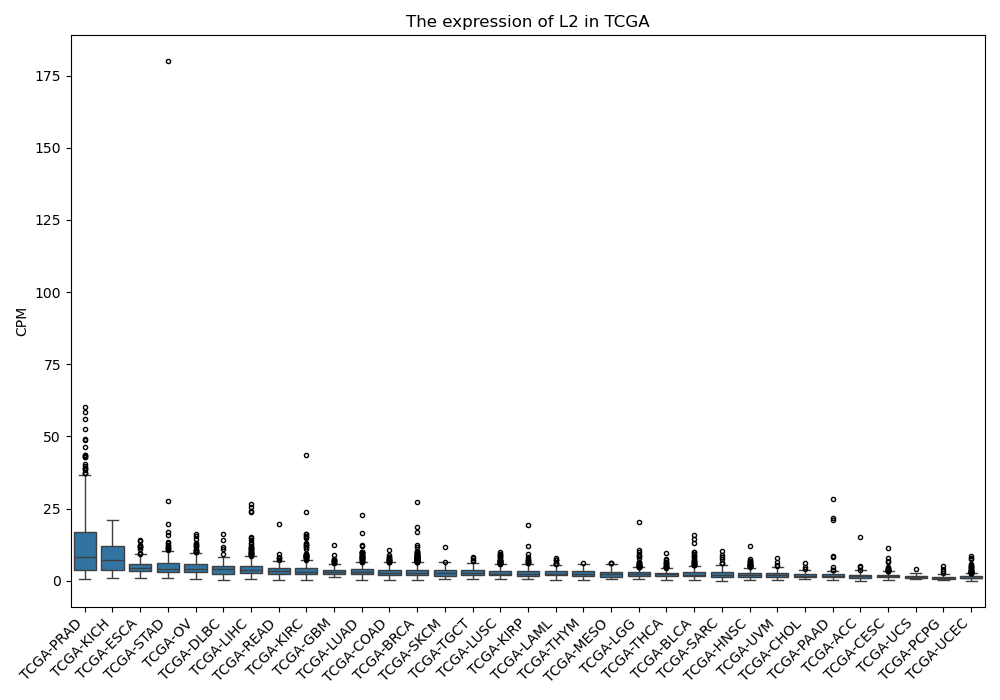
L2
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000358 |
|---|---|
| TE superfamily | L2 |
| TE class | LINE |
| Species | Mammalia |
| Length | 3082 |
| Kimura value | 37.63 |
| Tau index | 0.7930 |
| Description | L2 (LINE2) retrotransposon |
| Comment | L2 (LINE2) spread before the mammalian radiation, and copies are only 65-75% similar to the consensus. The 5' end is probably incomplete. The 3' end is also incomplete, but this terminus is extended by three model variants: L2a_3end, L2b_3end, and L2c_3end. Note that, whereas L1 is A- and purine-rich in the coding strand, L2 is C- and pyrimidine-rich. |
| Sequence |
ACCTCCNAGCCTCCATCGGCAGGGGTCAAGAGGGAGCCCCATGCGCTCNGAGTAAGACCGCCCTCCCCNGGCCTCCACGCCNCCATCGGCAGGAGTNAAGAGAGGGCCCCAGAACTCTGCGTGCTCCTGGCAAGACTCCNACTCCCCACTTCCCCCTCAACACTCACAGCCTCGCCACCACTATCTGCCTTGAGACCATCAGGACTAATCTCATCATGCCCTTTATCCCTGTTTACGTCAGTCACCACAAGAAGCCCAAGACTCCCAGGTCATCCCTCATCCCCCGGACTCTNATCCCCATACGGCTCTGTACATCTCCTNTTCTGACCCCATCTCCCACCCCTCCCCAACTCCTGAAACCCTTCCACTGTGCCCTCTGGAACTCACGGTCCGTCATCAGCAAAATCCCCCGTATCCTCAACCTCTTCTCTGAACGTTCCCTTCACCTTCTTGCTCTAACNGAAACCTGGCTCTCCCCTGAGGACACTGCTTCCCCTGCAGCCCTCTCAAGTGGTGGCTGTTTTCTCTCCCACANCCCTCGTACCACTGGGCCTGGAGGTGGGGTAGGTGTCCTCCTTGCTCCTCATTGCCGCTTCCAGACCATTCTCCCTCCCTCCTCCCTAAAACACCCCAGCTTTGAATCTCATGTCATCAGACTATACCACCCGCTACCCCTCCTTGTTGCAGTCATCTACCGACCTCCGGGTCACTCCCCCTCATTCCTTGAAGATTTTAGCTCCTGGCTCACTGTCACTCTCTCCAACACTACTCCTGTCATAATTCTTGGTGATTTCAATATCCACGTAGATGATCCTTCCAATACCCTGGCCTCTCAGTTCCTTGACCTCCTCTCCTCCAATGATCTTGTCCTCCACCCTACCTCAGCCACTCACTCCCATGGTCATACCCTAGACCTTGTCATTACCAATAACTGCAACCCCTCCATAATCTCAATTTCAAGCATCCCACTCTCTGACCACCACCTCCTATCTTTCCAGCTCACTCCCTCTAGTACCCCGACTCCAACAATTCTTCGACCCCACCGGGACCTCCAATCCATTGATCCTACCACCTTTTCACTGTCCCTCACCCCCCTCATGTCCTCACTTCCCTCCTTACCCAGCTTAGATTCCATGGTCNATCATTATAATCACTCCCTTGCATATACCCTCAACTCCCTTGCCCCTCTCTCGCTTCGTCGTACTCGCCTGGCAAAACCNCAACCCTGGTTAAATCCAACTCTCCGCCTACTCCGCGCCTGCACCCGNGCAGCTGAACGTGGCTGGAGAAAAACACACAACCATGCTGACTGGTCTCACTTTAAATTCATGACCACNAACCTCAAGTGGGCCCTTAATGCTGCCCGGCAATCNTACTACATTTCCCTAGTCCATTCACTCTCCCACTCTCCTAGACGACTATTTCACACCTTCTCCTCTCTCCTCAAACCTCCAACACCTCCTCCCCTATCCTCACTCTCAGCTGATGACCTTGCTTCCTATTTCACTGAGAAAATAGAAGCAATCAGAAGAGAACTTCCACANNCTCCCACCACCACATCTACCCACCTACCTGCATCTGTGCCCACATACTCTGCCTTCCCTCCTGTTACTACGGATGAACTGTCCGTGCTCCTATCTAAGGCCAACCCCTCCACTTGTGCACTAGATCCCATCCCCTCTCGCCTACTCAAGGACATCGCTCCAGCAATTCTCCCCTCTCTCTCCTGCATCATCAATTTTTCCCTCTCTACTGGATCATTCCCATCAGCATACAAACATGCTGTNATTTCTCCCATCTTTAAAAAACAAAAATTCTCCCTTGACCCCACNTCCCCCTCCAGCTACCGCCCCATTTCTCTGCTCCCCTTTACAGCAAAACTCCTCGAAAGAGTTGTCTATACTCGCTGTCTCCAATTCCTCTCCTCCCATTCTCTCTTGAACCCACTCCAATCAGGCTTTCGTCCCCACCACTCCACCGAAACTGCTCTTGTCAAGGTCACCAATGACCTCCACGTTGCTAAATCCAATGGTCAATTCTCAGTCCTCATCTTACTTGACCTNTCAGCAGCATTTGACACAGTTGATCACTCCCTCCTTCTTGAAACACTTTCTTCACTTGGCTTCCAGGACACCACACTCTCCTGGTTTTCCTCCTACCTCACTGGCCGCTCCTTCTCAGTCTCCTTTGCTGGTTCCTCCTCATCTCCCCGACCTCTNAACGTTGGAGTGCCCCAGGGCTCAGTCCTTGGACCTCTTCTCTTCTCTATCTACACTCACTCCCTNGGTGATCTCATCCAGTCTCATGGCTTTAAATACCATCTATATGCTGATGACTCCCAAATTTATATCTCCAGCCCGGACCTCTCCCCTGAACTCCAGACTCGTATATCCAACTGCCTACTCGACATCTCCACTTGGATGTCTAATAGGCATCTCAAACTTAACATGTCCAAAACTGAACTCCTGATCTTCCCCCTCAAACCTGCTCCTCCCGCAGTCTTCCCCATCTCAGTNAATGGCAACTCCATCCTTCCAGTTGCTCAGGCCAAAAACCTTGGAGTCATCCTTGACTCCTCTCTTTCTCTCACACCCCACATCCAATCCATCAGCAAATCCTGTCGGCTCTACCTTCAAAATATATCCAGAATCCGACCACTTCTCACCACCTCCACTGCCACCACCCTGGTCCAAGCCACCATCATCTCTCGCCTGGATTACTGCAATAGCCTCCTAACTGGTCTCCCTGCTTCCACCCTTGCCCCCCTNCAGTCTATTCTCAACACAGCAGCCAGAGTGATCCTTTTAAAACGTAAGTCAGATCATGTCACTCCTCTGCTCAAAACCCTCCAATGGCTTCCCATCTCACTCAGAGTAAAAGCCAAAGTCCTTACAATGGCCTACAAGGCCCTACATGATCTGGTCCCCCGTTACCTCTCTGACCTCATCTCCTACCACTCTCCCCCTCGCTCACTCCGCTCCAGCCACACTGGCCTCCTTGCTGTTCCTCGAACACGCCAGGCACGCTCCCGCCTCAGGGCCTTTGCACTTGCTGTTCCCTCTGCCTGGAACGCTCTTCCCCC
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| L2 | MYB17 | 1564 | 1574 | + | 16.28 | CCCACCTACCT |
| L2 | TBX3 | 1423 | 1431 | - | 16.16 | AGGTGTGAA |
| L2 | Nr5A2 | 1993 | 2001 | - | 16.13 | TGACCTTGA |
| L2 | ZNF189 | 2999 | 3007 | + | 16.13 | TGCTGTTCC |
| L2 | ZNF189 | 3050 | 3058 | + | 16.13 | TGCTGTTCC |
| L2 | TFAP2A | 2230 | 2240 | + | 16.09 | TGCCCCAGGGC |
| L2 | MAZ | 340 | 347 | + | 16.08 | CCCCTCCC |
| L2 | SP1 | 340 | 348 | - | 16.06 | GGGGAGGGG |
| L2 | LjSGA_053525.1 | 1346 | 1353 | - | 16.05 | GGGCCCAC |
| L2 | Prdm15 | 2142 | 2152 | - | 16.01 | GAAAACCAGGA |
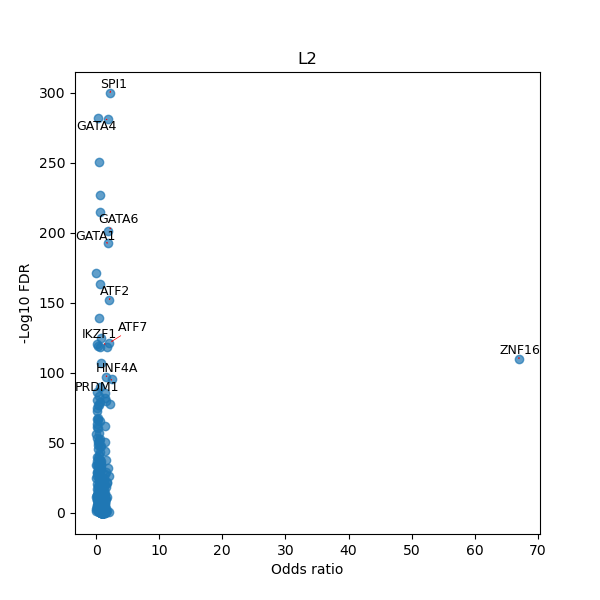
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.