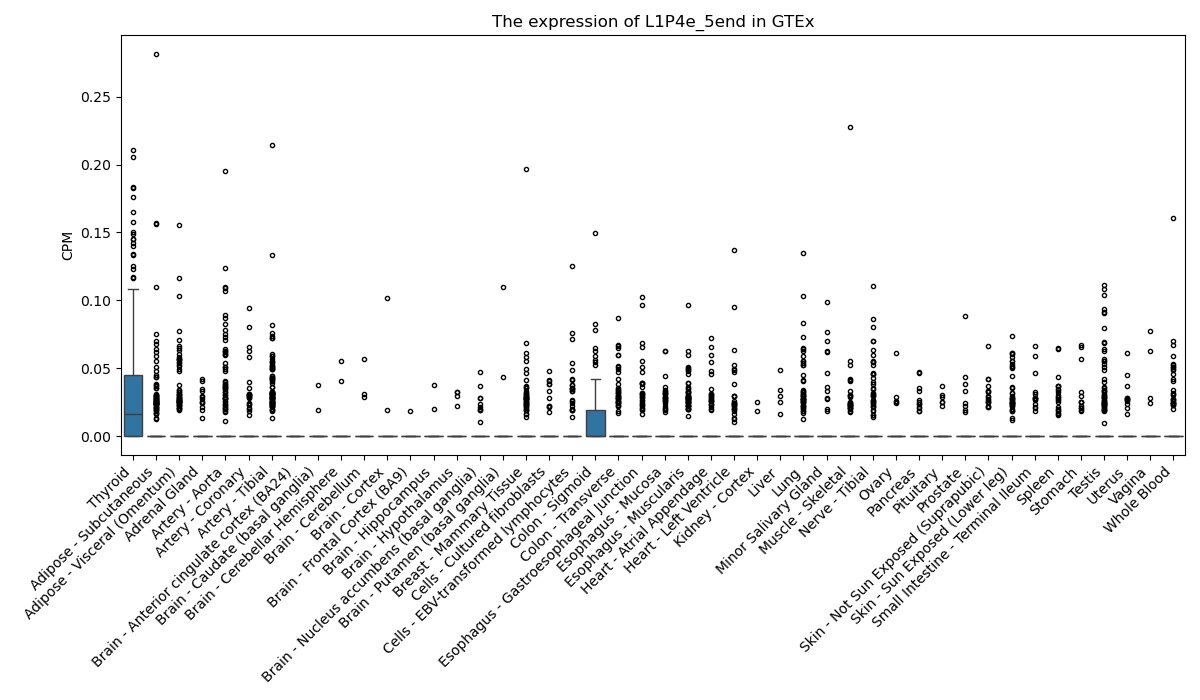
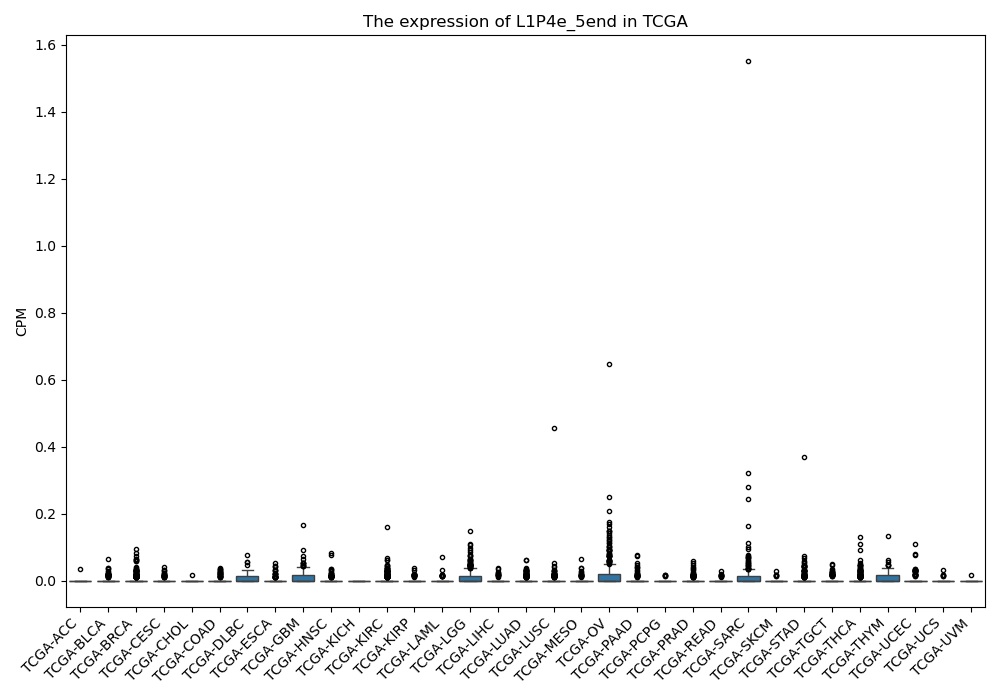
L1P4e_5end
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000325 |
|---|---|
| TE superfamily | L1 |
| TE class | LINE |
| Species | Haplorrhini |
| Length | 1391 |
| Kimura value | 17.78 |
| Tau index | 1.0000 |
| Description | 5' end of L1 retrotransposon, L1P4e_5end subfamily |
| Comment | - |
| Sequence |
GAGCCAAGATGGCCGACTAGACGCAGCCAGGAAGAGCTTCTCCCACCGAGAGAGACCAGACCATCAAGNAGACCGGCACACTCCGAGCAGATCTTCGGAAAGAAGGCATTGAGAGTGGACGGAGGGAGGACGCAGACCCCGGGCTGAAGGGGGAGGAAGCTGGGAACCCTGCACGGGGTTGCCGAGCACCAGGACTCGTTCCTGGCCCCGAGCGGCTCCTGGGGAAGGGGTGAGTNAAATAGGCGTGGAGTGGCCCACTCTCGCCACGGACCTCCGGAATCCTAGCTGCAGGAGACCCCACGACCCCCACGGACATTTGAGCTGGCAGGGAGAACTGCCCGGAGAGTTGGCAGAGACAGGACTCCAGCCTGCGCGGAGCCCAGAGGGTTTGGCGCGGGAACGGCTGCAGTGGAGCACGGCCANGGGCGCCCATCCCCCAAGGCTCGCCATACTCCTCTAGGTGGCTTTGGCCTTTGTTGGCTGCCGGACCTGGACAGAGCAGGGCTGTCTTGCCCGTGGGACTGGGGCCAGTCTGATCTGAGCGCCCCCCTGTCTGCCGGCCTCTCCCAGGGTCCCTGCCTGGCCGCACCCGCTTGCAGCGCAGCCTCAGNTGCCCAGCCGGGGCGCTTNCCAGCGGCCACCGCCATAGCTCTTTCGCCGGCAGACCCTCGCCNNCCCGTCGGAGNGCTTTTGCCGGTGCCCACGCACCCACCGCCGCCCTGCCGGCGCGCACTCGCCCGCAGCCTCCCCGCCGCCCTGCGGTGCGCATTGCCCACGGCCCCCACTGCCCCGCTGGCGCAATGCNTNCACACGGCGNCCCCCGCCGCCCCGCCGGAGCGCTTTTGCCGGCANCCCCCATCGGAGTGTTGTTGCCAGCGGACTGGGAACACNCTCGGCCCCTCCAGCGCAGCAGGTGCTTAACCTCGAGGGGCCAGAGAACAAAGCCGCGGGCCTGGTCCCAGCCCCCCAGGGTTAGAGCACGCAGCCCAGGAGTGCTGAGCTGAGCCTTGGCCCCCTGAAANCATCCAGAAACGAAGCCAGTCGACTAAACCCAACTTATACCACAGTCAAACCCTCAAGGGCATCAAAGAATATAAAAGCAAAAAGCCCCATCCAAAGGACAGCAACTTCAAAGATTAAAGGAACATCAGCCCACACAGATGAGAAAGAACCAGCGCAAGAACTCTGGCAACTCTAAAAGCCAGAGTGTCTTCTTACCTCCAAACGACCGCACTAGCTCCCCAGCAATGGTTCTTAACCAGACTGAAATGGCTGAAATGACAGACATAGAATTCAGAATCTGGATGGCAANGAAGNTCATCGAGATNCAGGAGAAGGTTGAAACCCAATCCAAGGAAANCAGTAAAACGATNCAAGAGCTGAAAGATGAC
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| L1P4e_5end | Zm00001d002364 | 747 | 756 | + | 16.53 | CCCCGCCGCC |
| L1P4e_5end | Zm00001d002364 | 819 | 828 | + | 16.53 | CCCCGCCGCC |
| L1P4e_5end | USV1 | 1011 | 1020 | + | 16.51 | GCCCCCTGAA |
| L1P4e_5end | Zm00001d052229 | 747 | 756 | + | 16.50 | CCCCGCCGCC |
| L1P4e_5end | Zm00001d052229 | 819 | 828 | + | 16.50 | CCCCGCCGCC |
| L1P4e_5end | DREB2D | 706 | 719 | + | 16.45 | CACCCACCGCCGCC |
| L1P4e_5end | PATZ1 | 818 | 828 | - | 16.36 | GGCGGCGGGGG |
| L1P4e_5end | Zm00001d049364 | 709 | 719 | + | 16.22 | CCACCGCCGCC |
| L1P4e_5end | LEP | 710 | 719 | + | 16.20 | CACCGCCGCC |
| L1P4e_5end | ERF087 | 710 | 719 | - | 16.14 | GGCGGCGGTG |
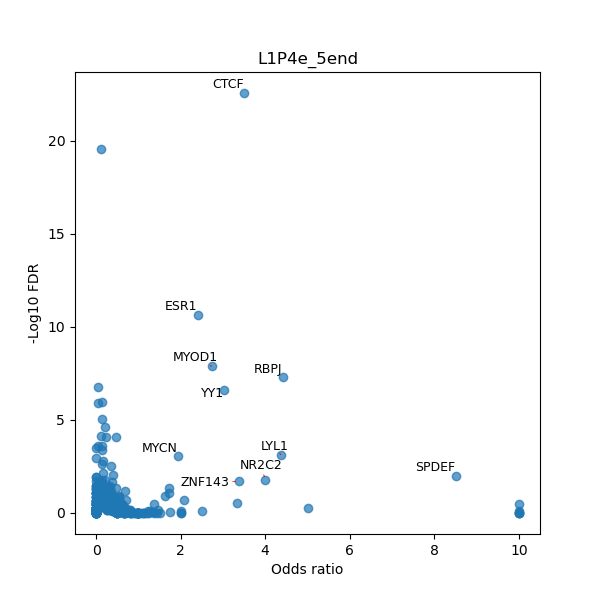
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.