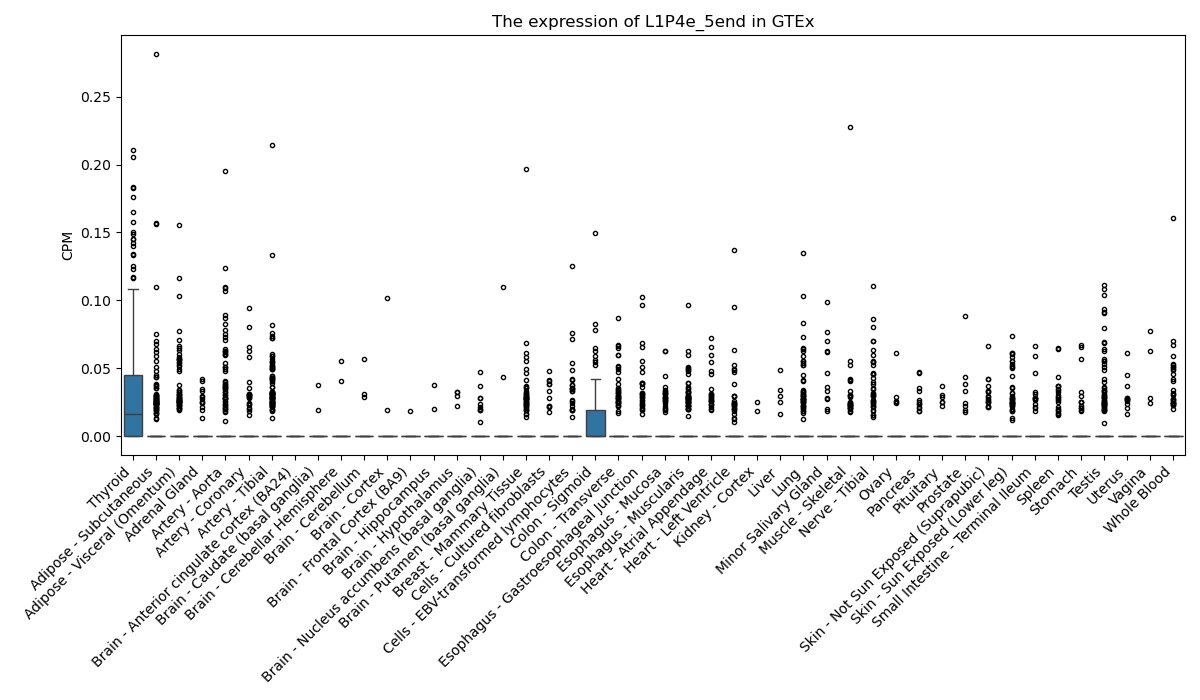
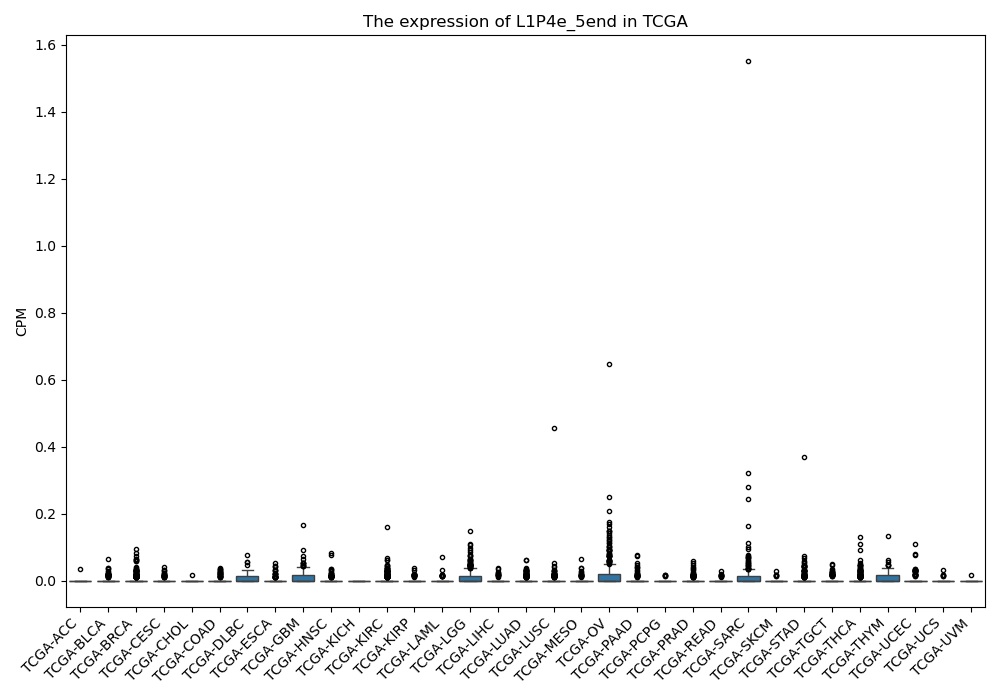
L1P4e_5end
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000325 |
|---|---|
| TE superfamily | L1 |
| TE class | LINE |
| Species | Haplorrhini |
| Length | 1391 |
| Kimura value | 17.78 |
| Tau index | 1.0000 |
| Description | 5' end of L1 retrotransposon, L1P4e_5end subfamily |
| Comment | - |
| Sequence |
GAGCCAAGATGGCCGACTAGACGCAGCCAGGAAGAGCTTCTCCCACCGAGAGAGACCAGACCATCAAGNAGACCGGCACACTCCGAGCAGATCTTCGGAAAGAAGGCATTGAGAGTGGACGGAGGGAGGACGCAGACCCCGGGCTGAAGGGGGAGGAAGCTGGGAACCCTGCACGGGGTTGCCGAGCACCAGGACTCGTTCCTGGCCCCGAGCGGCTCCTGGGGAAGGGGTGAGTNAAATAGGCGTGGAGTGGCCCACTCTCGCCACGGACCTCCGGAATCCTAGCTGCAGGAGACCCCACGACCCCCACGGACATTTGAGCTGGCAGGGAGAACTGCCCGGAGAGTTGGCAGAGACAGGACTCCAGCCTGCGCGGAGCCCAGAGGGTTTGGCGCGGGAACGGCTGCAGTGGAGCACGGCCANGGGCGCCCATCCCCCAAGGCTCGCCATACTCCTCTAGGTGGCTTTGGCCTTTGTTGGCTGCCGGACCTGGACAGAGCAGGGCTGTCTTGCCCGTGGGACTGGGGCCAGTCTGATCTGAGCGCCCCCCTGTCTGCCGGCCTCTCCCAGGGTCCCTGCCTGGCCGCACCCGCTTGCAGCGCAGCCTCAGNTGCCCAGCCGGGGCGCTTNCCAGCGGCCACCGCCATAGCTCTTTCGCCGGCAGACCCTCGCCNNCCCGTCGGAGNGCTTTTGCCGGTGCCCACGCACCCACCGCCGCCCTGCCGGCGCGCACTCGCCCGCAGCCTCCCCGCCGCCCTGCGGTGCGCATTGCCCACGGCCCCCACTGCCCCGCTGGCGCAATGCNTNCACACGGCGNCCCCCGCCGCCCCGCCGGAGCGCTTTTGCCGGCANCCCCCATCGGAGTGTTGTTGCCAGCGGACTGGGAACACNCTCGGCCCCTCCAGCGCAGCAGGTGCTTAACCTCGAGGGGCCAGAGAACAAAGCCGCGGGCCTGGTCCCAGCCCCCCAGGGTTAGAGCACGCAGCCCAGGAGTGCTGAGCTGAGCCTTGGCCCCCTGAAANCATCCAGAAACGAAGCCAGTCGACTAAACCCAACTTATACCACAGTCAAACCCTCAAGGGCATCAAAGAATATAAAAGCAAAAAGCCCCATCCAAAGGACAGCAACTTCAAAGATTAAAGGAACATCAGCCCACACAGATGAGAAAGAACCAGCGCAAGAACTCTGGCAACTCTAAAAGCCAGAGTGTCTTCTTACCTCCAAACGACCGCACTAGCTCCCCAGCAATGGTTCTTAACCAGACTGAAATGGCTGAAATGACAGACATAGAATTCAGAATCTGGATGGCAANGAAGNTCATCGAGATNCAGGAGAAGGTTGAAACCCAATCCAAGGAAANCAGTAAAACGATNCAAGAGCTGAAAGATGAC
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| L1P4e_5end | KLF17 | 700 | 713 | + | 19.19 | CCCACGCACCCACC |
| L1P4e_5end | ERF057 | 706 | 719 | + | 18.10 | CACCCACCGCCGCC |
| L1P4e_5end | DREB2F | 709 | 719 | + | 17.65 | CCACCGCCGCC |
| L1P4e_5end | EREB29 | 748 | 757 | + | 17.61 | CCCGCCGCCC |
| L1P4e_5end | EREB29 | 820 | 829 | + | 17.61 | CCCGCCGCCC |
| L1P4e_5end | ZNF75A | 36 | 47 | + | 17.12 | GCTTCTCCCACC |
| L1P4e_5end | Zm00001d049364 | 818 | 828 | + | 17.04 | CCCCCGCCGCC |
| L1P4e_5end | ZNF281 | 429 | 438 | - | 16.91 | GGGGGATGGG |
| L1P4e_5end | ERF014 | 632 | 646 | + | 16.73 | CAGCGGCCACCGCCA |
| L1P4e_5end | EREB29 | 711 | 720 | + | 16.70 | ACCGCCGCCC |
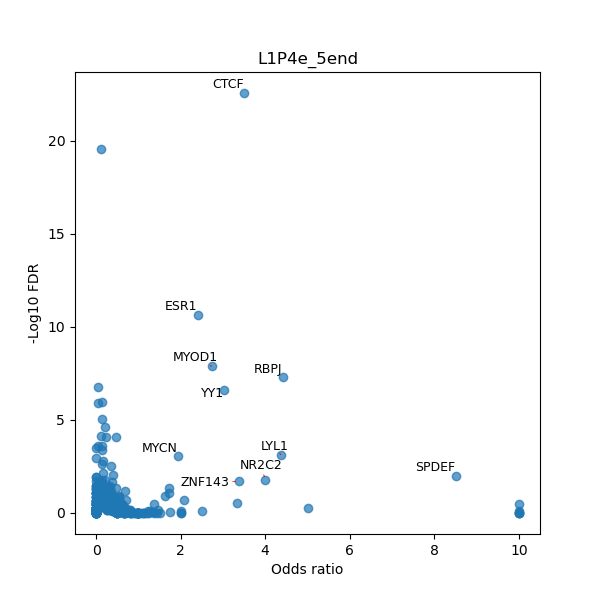
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.