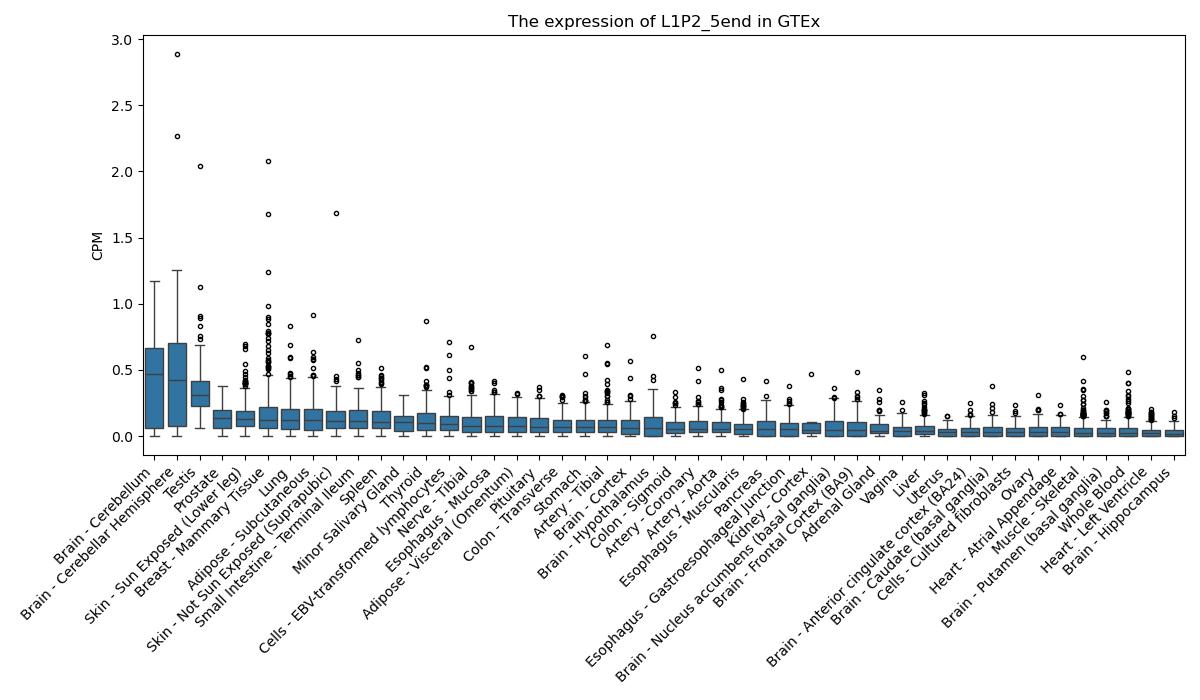
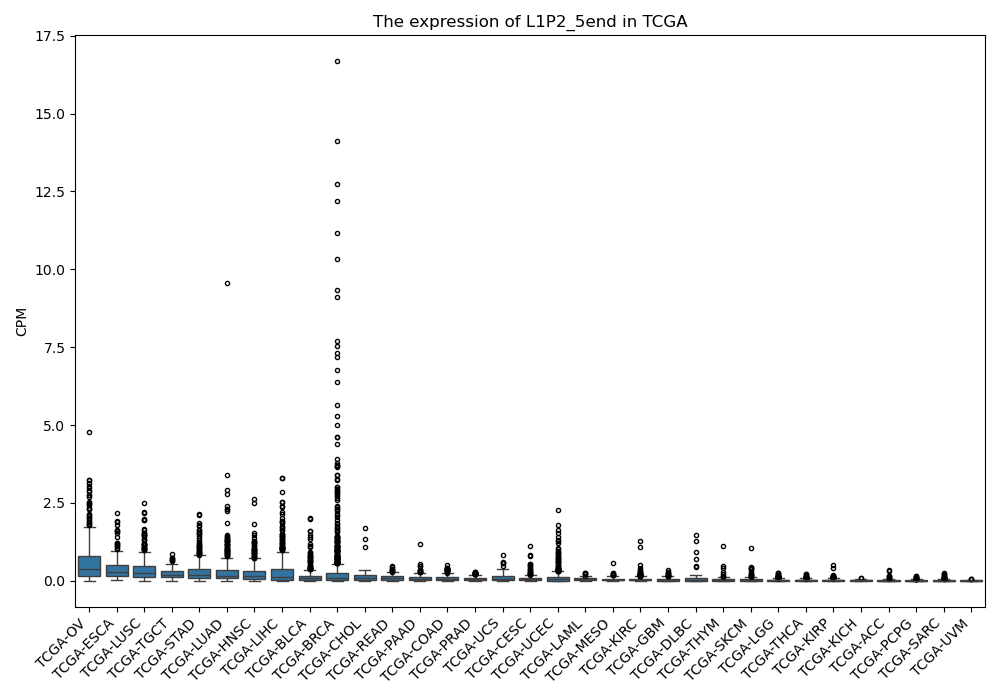
L1P2_5end
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000317 |
|---|---|
| TE superfamily | L1 |
| TE class | LINE |
| Species | Catarrhini |
| Length | 2275 |
| Kimura value | 5.44 |
| Tau index | 0.8359 |
| Description | 5' end of L1 retrotransposon, L1P2_5end subfamily |
| Comment | - |
| Sequence |
TCCGTTCCAAGATGGCCGAATAGGAACAGCTCCGGTCTGCAGCTCCCAGCGTGATCGACGCAGAAGACGGGTGATTTCTGCATTTCCAACTGAGGTACCTGGTTCATCTCACTGGGACTGGTTGGACAGTGGGTGCAGCCCACGGAGGGCGAGCCGAAGCAGGGCGGGGCATCGCCTCACCCGGGAAGCGCAAGGGGTCGGGGGATTTCCCTTTCCTAGCCAAGGGAAGCCGTGACAGACTGTACCTGGAAAANCGGGACACTCCCGCCCAAATACTGCGCTTTTCCAACGGTCTTAGCAAACGGCACACCAGGAGATTNTATCCCGCGCCTGGCTCGGCGGGTCCCACGCCCACGGAGCCTTGCTCACTGCTAGCGCAGCAGTCTGAGATCGANCTGCGAGGCGGCAGCCTGGCTGGGGGAGGGGCGTCCGCCATTGCTGAGGCTTGAGTAGGTAAACAAAGCGGCCGGGAAGCTCGAACTGGGCGGAGCCCACCGCAGCTCAGCGAGGCCTGCTGCCTCTGTAGACTCCACCTCTGGGGGCAGGGCATAGCTGAACAAAAGGCAGCAGACAACTTCTGCAGACTTAAACGTCCCTGTCTGACAGCTCTGAAGAGAGCAGTGGTTCTCCCAGCACGGCGTTTGAGCTCTGAGAACGGACAGACTGCCTCCTCAAGTGGGTCCCTGACCCCCGTGTAGCCTAACTGGGAGACACCTCCCAGTAGGGGCCGACNGACACCTCATACAGGCGGGTGCCCCTCTGGGACGAAGCTTCCAGAGGAAGGATCAGGCAGCAATATTTGCTGTTCTGCAATATTTGCTGTTCTGCAGCCTCCGCTGGTGATACCCAGGCAAACAGGGTCTGGAGTGGACCTCCAGCAAACTCCAACAGACCTGCAGCTGAGGGNCCTGACTGTTAGAAGGAAAACTAACAAACAGAAAGGAATAGCATCAACATCAACAAAAAGGACATCCACACCAAAACCCCATCTGTAGGTCACCANCATCAAAGACCAAAGGTAGATAAAACCACAAAGATGGGGAGAAACCAGAGCAGAAAAGCTGAAAATTCTAAAAACCAGAGCGCCTCTTCTCCTCCAAAGGATCGCAGCTCCTCGCCAGCAACGGAACAAAGCTGGACGGAGAATGACTTTGACGAGTTGACAGAAGTAGGCTTCAGAAGGTCGGTAATAACAAACTTCTCCGAGCTAAAGGAGGATGTTCGAACCCATCGCAAGGAAGCTAAAAACCTTGAAAAAAGATTAGACGAATGGCTAACTAGAATAAACAGTGTAGAGAAGACCTTAAATGACCTGATGGAGCTGAAAACCACGGCACGAGAACTNCGTGACGCATGCACAAGCTTCAGTAGCCGATTCGATCAAGTGGAAGAAAGGGTATCAGTGATTGAAGATCAAATTAATGAAATAAAGCGAGAAGANAAGNTTAGAGAAAAAAGAGTAAAAAGAAACGAACAAAGCCTCCAAGAAATATGGGACTATGTGAAAAGACCAAATCTACGTTTGATTGGTGTACCTGAAAGTGACGGGGAGAATGGAACCAAGTTGGAAAACACTCTTCAGGATATTATCCAGGAGAACTTCCCCAACCTAGCAAGGCAGGCCAACATTCAAATTCAGGAAATACAGAGAACGCCACAAAGATACTCCTCGAGAAGAGCAACCCCAAGACACATAATTGTCAGATTCACCAAGGTTGAAATGAAGGAAAAAATGTTAAGGGCAGCCAGAGAGAAAGGTCGGGTTACCCACAAAGGGAAGCCCATCAGACTAACAGCGGATCTCTCGGCAGAAACCCTACAAGCCAGAAGAGAGTGGGGGCCAATATTCAACATTCTTAAAGAAAAGAATTTTCAACCCAGAATTTCATATCCAGCCAAACTAAGCTTCATAAGTGAAGGAGAAATAAAATCCTTTACAGACAAGCAAATGCTGAGAGATTTTGTCACCACCAGGCCTGCCTTACAAGAGCTCCTGAAGGAAGCACTAAACATGGAAAGGAACAACCGGTACCAGCCACTGCAAAAACATGCCAAATTGTAAAGACCATCGATGCTAGGAAGAAACTGCATCAACTAACGGGCAAAATAACCAGCTAACATCATAATGACAGGATCAAATTCACACATAACAATATTAACCTTAAATGTAAATGGGCTAAATGCCCCAATTAAAAGACACAGACTGGCAAATTGGATAAAGAGTCAAGACCCATCAGTGTGCTGTATTCAGGAGACCCATCTCACGTGCAGAGAC
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| L1P2_5end | BPC5 | 1639 | 1668 | + | -45.00 | GGAAATACAGAGAACGCCACAAAGATACTC |
| L1P2_5end | BPC5 | 1726 | 1755 | + | -47.05 | GGAAAAAATGTTAAGGGCAGCCAGAGAGAA |
| L1P2_5end | BPC5 | 1021 | 1050 | + | -47.37 | AGATAAAACCACAAAGATGGGGAGAAACCA |
| L1P2_5end | DAL81 | 321 | 339 | - | -52.00 | CCGAGCCAGGCGCGGGATA |
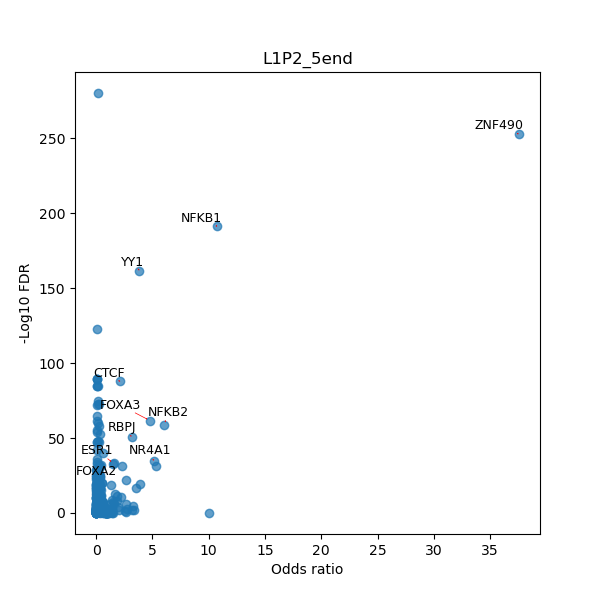
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.