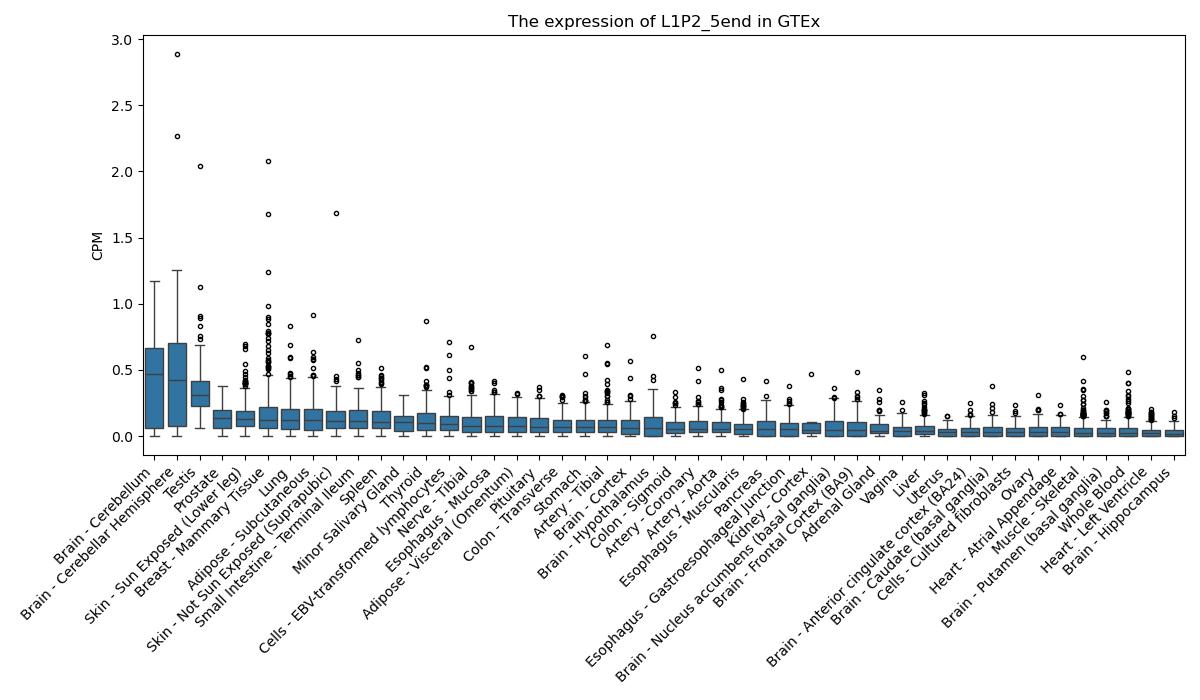
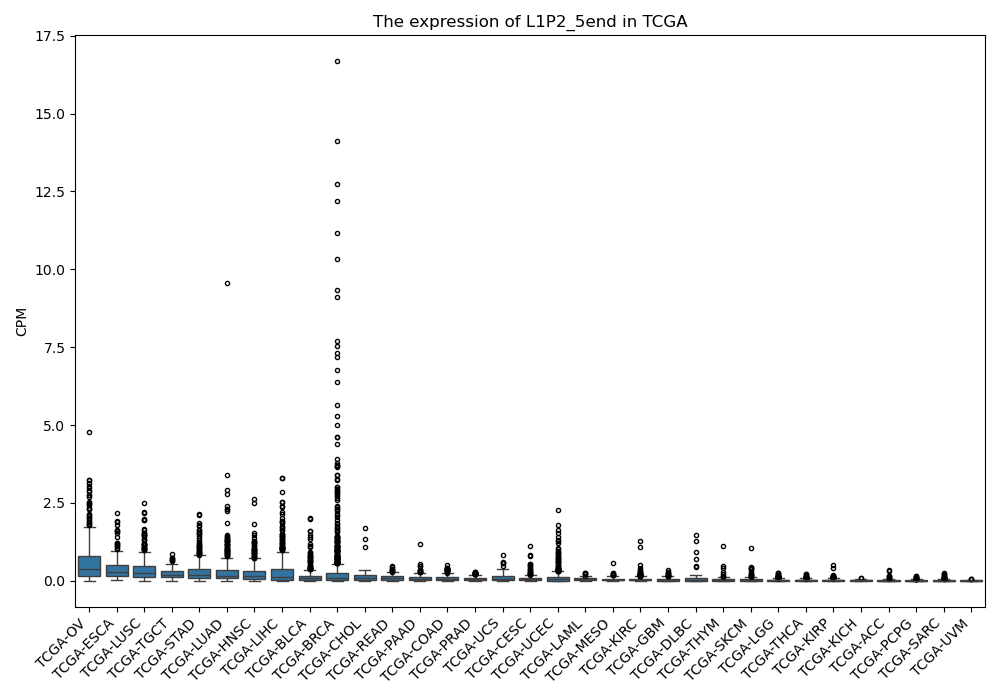
L1P2_5end
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000317 |
|---|---|
| TE superfamily | L1 |
| TE class | LINE |
| Species | Catarrhini |
| Length | 2275 |
| Kimura value | 5.44 |
| Tau index | 0.8359 |
| Description | 5' end of L1 retrotransposon, L1P2_5end subfamily |
| Comment | - |
| Sequence |
TCCGTTCCAAGATGGCCGAATAGGAACAGCTCCGGTCTGCAGCTCCCAGCGTGATCGACGCAGAAGACGGGTGATTTCTGCATTTCCAACTGAGGTACCTGGTTCATCTCACTGGGACTGGTTGGACAGTGGGTGCAGCCCACGGAGGGCGAGCCGAAGCAGGGCGGGGCATCGCCTCACCCGGGAAGCGCAAGGGGTCGGGGGATTTCCCTTTCCTAGCCAAGGGAAGCCGTGACAGACTGTACCTGGAAAANCGGGACACTCCCGCCCAAATACTGCGCTTTTCCAACGGTCTTAGCAAACGGCACACCAGGAGATTNTATCCCGCGCCTGGCTCGGCGGGTCCCACGCCCACGGAGCCTTGCTCACTGCTAGCGCAGCAGTCTGAGATCGANCTGCGAGGCGGCAGCCTGGCTGGGGGAGGGGCGTCCGCCATTGCTGAGGCTTGAGTAGGTAAACAAAGCGGCCGGGAAGCTCGAACTGGGCGGAGCCCACCGCAGCTCAGCGAGGCCTGCTGCCTCTGTAGACTCCACCTCTGGGGGCAGGGCATAGCTGAACAAAAGGCAGCAGACAACTTCTGCAGACTTAAACGTCCCTGTCTGACAGCTCTGAAGAGAGCAGTGGTTCTCCCAGCACGGCGTTTGAGCTCTGAGAACGGACAGACTGCCTCCTCAAGTGGGTCCCTGACCCCCGTGTAGCCTAACTGGGAGACACCTCCCAGTAGGGGCCGACNGACACCTCATACAGGCGGGTGCCCCTCTGGGACGAAGCTTCCAGAGGAAGGATCAGGCAGCAATATTTGCTGTTCTGCAATATTTGCTGTTCTGCAGCCTCCGCTGGTGATACCCAGGCAAACAGGGTCTGGAGTGGACCTCCAGCAAACTCCAACAGACCTGCAGCTGAGGGNCCTGACTGTTAGAAGGAAAACTAACAAACAGAAAGGAATAGCATCAACATCAACAAAAAGGACATCCACACCAAAACCCCATCTGTAGGTCACCANCATCAAAGACCAAAGGTAGATAAAACCACAAAGATGGGGAGAAACCAGAGCAGAAAAGCTGAAAATTCTAAAAACCAGAGCGCCTCTTCTCCTCCAAAGGATCGCAGCTCCTCGCCAGCAACGGAACAAAGCTGGACGGAGAATGACTTTGACGAGTTGACAGAAGTAGGCTTCAGAAGGTCGGTAATAACAAACTTCTCCGAGCTAAAGGAGGATGTTCGAACCCATCGCAAGGAAGCTAAAAACCTTGAAAAAAGATTAGACGAATGGCTAACTAGAATAAACAGTGTAGAGAAGACCTTAAATGACCTGATGGAGCTGAAAACCACGGCACGAGAACTNCGTGACGCATGCACAAGCTTCAGTAGCCGATTCGATCAAGTGGAAGAAAGGGTATCAGTGATTGAAGATCAAATTAATGAAATAAAGCGAGAAGANAAGNTTAGAGAAAAAAGAGTAAAAAGAAACGAACAAAGCCTCCAAGAAATATGGGACTATGTGAAAAGACCAAATCTACGTTTGATTGGTGTACCTGAAAGTGACGGGGAGAATGGAACCAAGTTGGAAAACACTCTTCAGGATATTATCCAGGAGAACTTCCCCAACCTAGCAAGGCAGGCCAACATTCAAATTCAGGAAATACAGAGAACGCCACAAAGATACTCCTCGAGAAGAGCAACCCCAAGACACATAATTGTCAGATTCACCAAGGTTGAAATGAAGGAAAAAATGTTAAGGGCAGCCAGAGAGAAAGGTCGGGTTACCCACAAAGGGAAGCCCATCAGACTAACAGCGGATCTCTCGGCAGAAACCCTACAAGCCAGAAGAGAGTGGGGGCCAATATTCAACATTCTTAAAGAAAAGAATTTTCAACCCAGAATTTCATATCCAGCCAAACTAAGCTTCATAAGTGAAGGAGAAATAAAATCCTTTACAGACAAGCAAATGCTGAGAGATTTTGTCACCACCAGGCCTGCCTTACAAGAGCTCCTGAAGGAAGCACTAAACATGGAAAGGAACAACCGGTACCAGCCACTGCAAAAACATGCCAAATTGTAAAGACCATCGATGCTAGGAAGAAACTGCATCAACTAACGGGCAAAATAACCAGCTAACATCATAATGACAGGATCAAATTCACACATAACAATATTAACCTTAAATGTAAATGGGCTAAATGCCCCAATTAAAAGACACAGACTGGCAAATTGGATAAAGAGTCAAGACCCATCAGTGTGCTGTATTCAGGAGACCCATCTCACGTGCAGAGAC
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| L1P2_5end | ZNF354A | 2163 | 2182 | + | 20.38 | TAAATGTAAATGGGCTAAAT |
| L1P2_5end | ZNF281 | 417 | 426 | + | 20.07 | GGGGGAGGGG |
| L1P2_5end | ZNF136 | 1872 | 1886 | - | 19.65 | AAATTCTGGGTTGAA |
| L1P2_5end | DOF3.6 | 1451 | 1471 | - | 19.12 | GTTTCTTTTTACTCTTTTTTC |
| L1P2_5end | ZNF148 | 417 | 426 | - | 18.97 | CCCCTCCCCC |
| L1P2_5end | PATZ1 | 417 | 427 | + | 18.42 | GGGGGAGGGGC |
| L1P2_5end | KLF11 | 346 | 355 | + | 17.79 | CCACGCCCAC |
| L1P2_5end | NFKB2 | 201 | 211 | - | 17.53 | GGGAAATCCCC |
| L1P2_5end | NFKB2 | 201 | 211 | + | 17.47 | GGGGATTTCCC |
| L1P2_5end | Dif | 202 | 211 | + | 17.39 | GGGATTTCCC |
TFBS enrichment in GRCh38
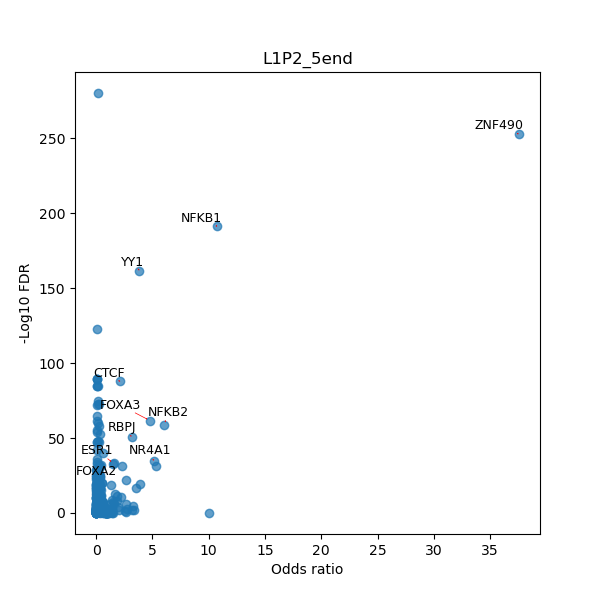
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.