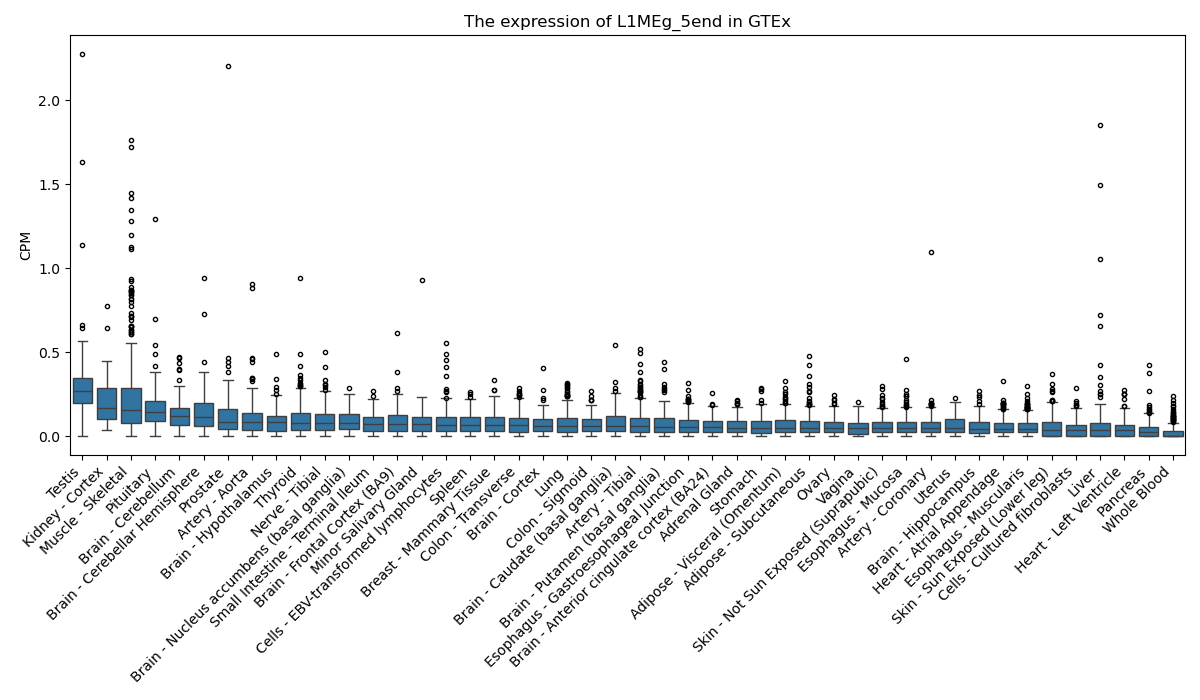
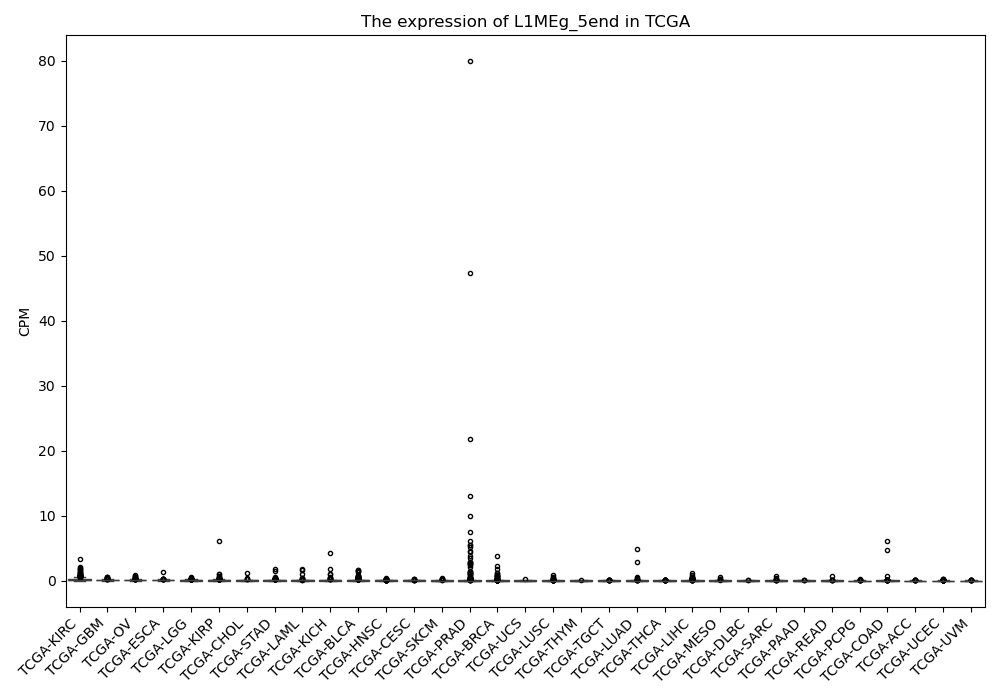
L1MEg_5end
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000311 |
|---|---|
| TE superfamily | L1 |
| TE class | LINE |
| Species | Theria_mammals |
| Length | 2101 |
| Kimura value | 29.58 |
| Tau index | 0.7568 |
| Description | 5' end of L1 retrotransposon, L1MEg_5end subfamily |
| Comment | - |
| Sequence |
TTCCGCTTCCGGCAATGGCGGACTAGNTTGTTCAGACCAACCCTCCCGCTGAGAACAACTAGAAAAGCTGGACAAAATATAAAAAACATCTGTTTGAAGGCATCGGAGAGCTACCAAGGCAGCGAGGAATTTGAGGGGCCAAGATCCCGGAGAGAAGGGAAGCCCAGAGAGGTGAGCCCGACATTCGGNGCCGCTTTTCCCCTCGAGGCATTTGCCGATTCCGAAAGCGGCGGCTGAGAGGCTGAGAAGCTGAGCAGAGCTTTCGGCAGTCTCACGGGGCTGGGGAGACAAAAATTGGAGTTCAGGGCCCGCCAAGGAGGAGGGGCCCTGGTAAACCCCAGCTTTCAGTTGGGACCCCGAAGGGCTACACCCTAGGAGTAAGGGCGAACCGGAAATAGACCAGCCCTCACAAAGACTGAAGCCCAGCTTCGAATCANCTCAATCCCTGATTGGATTAAGGTGATCTGATTGCTAGTGCCCCTAGCTGCCTGCCAGAAGCAAAAGTAAATCCTCTCTGGAGGAAGATAACATCATCCAGAGCCTCAAATTATCTCTACAATTTTTCATATACAATGTCCGGCATTCAATCAAAAATNACCAGGCATACNAGGAGACAAGACAAATGACCGAAAANCAAGAGAAAAACAGACAATAGAAACAGACCCACAGGNGATCCAGATATTGGAGTTATCAGACACGGACTTTAAAATAACTATGATTAATATGTTCAAGAAAATAAANGACAAGATGGAGAATTTCAGCAGAGAACTGGAANCTATAAAAAAGAATCAAATGGAAATTCTAGAACTGAAAAATATACAATAACTGAAATTAAGAACTCAATAGATGGGTTTAACAGCAGATTAGACACAGCNGAAGAGAGGATTAGTGAACTGGAAGATAGGTCAGNAGAAAATATCCAGACTGAAGCACAGAGAGANAAAAAAATGGAAAATACAGAAAAGAGCGTAAGAGACATATGGGACACGGTGAAAAGGTCTAACATACGTGTAATTGGAGTCCCAGAAGGAGAGGAGAGAGAGAATGGGGCAGAAGCAATATTTGAAGAGATAATGGCCGAGAATTTTCCAAAACTGATGAAAGACATCAANCCACAGATTCAAGAAGCTCTACGAACCCCAAGCAGGATAAATACAAAGAAAACCACACCTAGGCACATCATAGTAAAACTGCTGAAAACCAAAGACAAAGAGAAAATCTTAAAAGCAGCCAGAGAAAAAAGACACATTACCTTCAAAGGAGCAACAATAAGACTGACAGCTGACTTCTCAACAGAAACAATGGAAGCCAGAAGACAATGGAATGACATCTTTAAAGTGCTGAAAGAAAATAACTGCCAACCTAGAATTCTATACCCAGGAAAATATCCTTCAAAAATGAAGGCGAAATAAAGACATTTTCAGACAAACAAAAACTGAGAGAATTCGTCACCAGCAGACCTGCACTAAAAGAAATACTAAAGGAAGTTCTTCAGGCAGAAGGAAAATGATCCCAGATGGAAGCACGGAAATGCAGGAAGGAATGAAGAGCAACGGAAAGGGTAAATATGTGGGTAAATCTAAATGAATATTGACTGTATAAAACAATAATAATAATGTCTTGTGGGGTTTAAAATATATATAGAATTAAAATACATGACAACAATAGCACAAAAGGGAAGAGAGGGTAAATGGAGTTAAAGTGTTCTAAGGTCCTTGCATTGTCCGGGAAGTGGTAAAGTACTAATTTATATTAGACTTTAATAAGTCAAGGATGCATGTTGTAATCTCTAGGGTAACCACTAAAAGAATAGTAAAAGAATGTATAACTAACAAGCTAATAGAGAGGGAAAATGGAATAATAAAAATATTCGATTAATCCAAAAGAAGGCAAGAAAAAAGGAAAAAANAGAAAGAAAAACAGATGGGACAAATAGAAAACAAATAGTAAGATGGTAGATTTAAACCCAAATATATCAGTAATTACATTAAATGTAAATGGACTAAATACTCCAATTNAAAGANAAAGATTGTCAGACTGGATTTAAAAACAAAACCCAACTATATGCTGCTTACAAGAGACACACCTTAAATATAAGGAC
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| L1MEg_5end | TFAP2C | 198 | 210 | - | 15.07 | TGCCTCGAGGGGA |
| L1MEg_5end | PI | 942 | 955 | + | 15.05 | AAAAAAATGGAAAA |
| L1MEg_5end | FEV | 388 | 396 | + | 15.02 | ACCGGAAAT |
| L1MEg_5end | ZNF281 | 316 | 325 | + | 15.01 | GGAGGAGGGG |
| L1MEg_5end | TFAP2A | 198 | 210 | + | 14.98 | TCCCCTCGAGGCA |
| L1MEg_5end | TFAP2A | 198 | 210 | - | 14.98 | TGCCTCGAGGGGA |
| L1MEg_5end | ZBTB7A | 4 | 12 | - | 14.98 | CCGGAAGCG |
| L1MEg_5end | eor-1 | 1203 | 1215 | + | 14.85 | AAAGACAAAGAGA |
| L1MEg_5end | GT-4 | 710 | 721 | + | 14.83 | TAACTATGATTA |
| L1MEg_5end | PHOX2A | 1980 | 1990 | - | 14.82 | TAATGTAATTA |
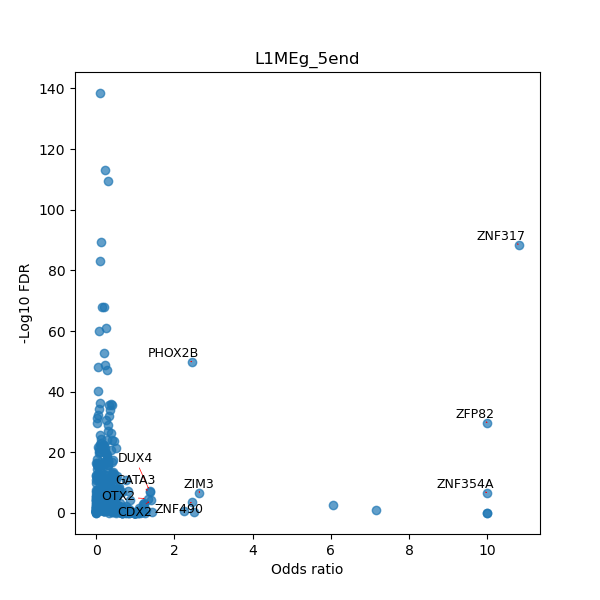
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.