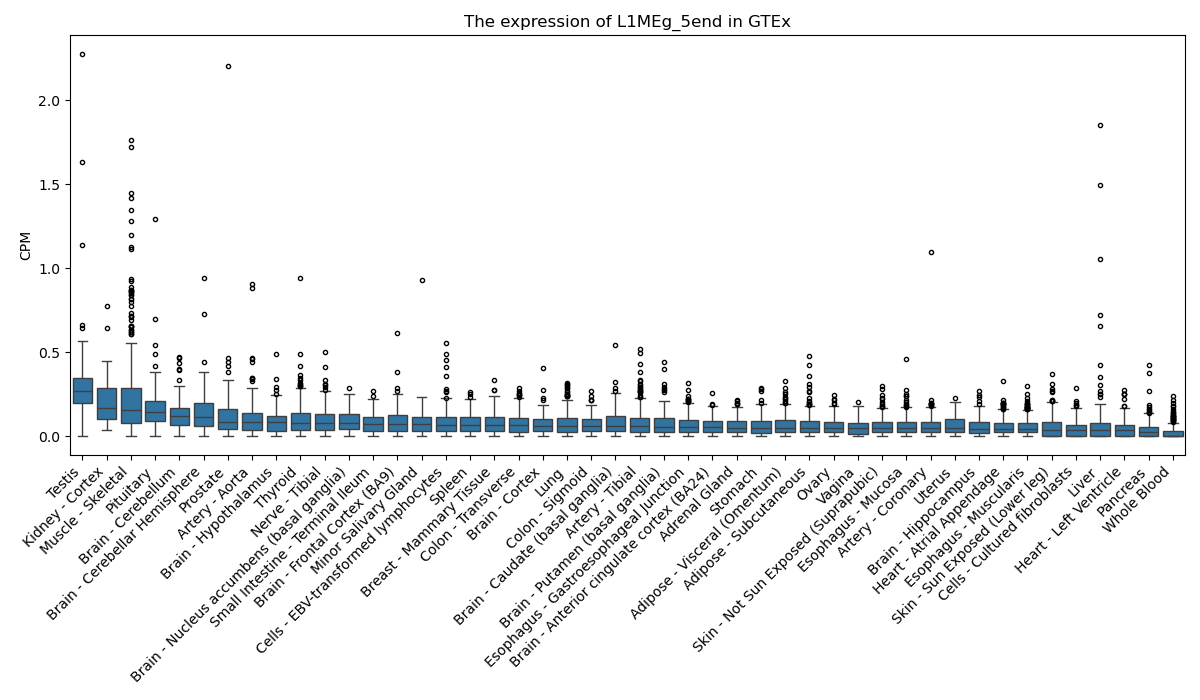
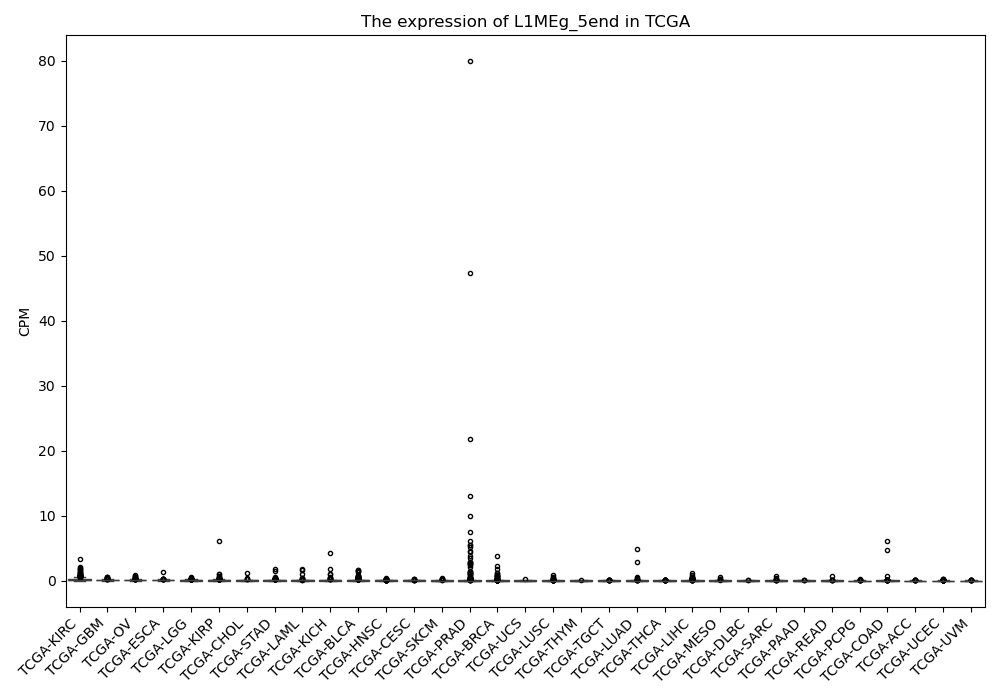
L1MEg_5end
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000311 |
|---|---|
| TE superfamily | L1 |
| TE class | LINE |
| Species | Theria_mammals |
| Length | 2101 |
| Kimura value | 29.58 |
| Tau index | 0.7568 |
| Description | 5' end of L1 retrotransposon, L1MEg_5end subfamily |
| Comment | - |
| Sequence |
TTCCGCTTCCGGCAATGGCGGACTAGNTTGTTCAGACCAACCCTCCCGCTGAGAACAACTAGAAAAGCTGGACAAAATATAAAAAACATCTGTTTGAAGGCATCGGAGAGCTACCAAGGCAGCGAGGAATTTGAGGGGCCAAGATCCCGGAGAGAAGGGAAGCCCAGAGAGGTGAGCCCGACATTCGGNGCCGCTTTTCCCCTCGAGGCATTTGCCGATTCCGAAAGCGGCGGCTGAGAGGCTGAGAAGCTGAGCAGAGCTTTCGGCAGTCTCACGGGGCTGGGGAGACAAAAATTGGAGTTCAGGGCCCGCCAAGGAGGAGGGGCCCTGGTAAACCCCAGCTTTCAGTTGGGACCCCGAAGGGCTACACCCTAGGAGTAAGGGCGAACCGGAAATAGACCAGCCCTCACAAAGACTGAAGCCCAGCTTCGAATCANCTCAATCCCTGATTGGATTAAGGTGATCTGATTGCTAGTGCCCCTAGCTGCCTGCCAGAAGCAAAAGTAAATCCTCTCTGGAGGAAGATAACATCATCCAGAGCCTCAAATTATCTCTACAATTTTTCATATACAATGTCCGGCATTCAATCAAAAATNACCAGGCATACNAGGAGACAAGACAAATGACCGAAAANCAAGAGAAAAACAGACAATAGAAACAGACCCACAGGNGATCCAGATATTGGAGTTATCAGACACGGACTTTAAAATAACTATGATTAATATGTTCAAGAAAATAAANGACAAGATGGAGAATTTCAGCAGAGAACTGGAANCTATAAAAAAGAATCAAATGGAAATTCTAGAACTGAAAAATATACAATAACTGAAATTAAGAACTCAATAGATGGGTTTAACAGCAGATTAGACACAGCNGAAGAGAGGATTAGTGAACTGGAAGATAGGTCAGNAGAAAATATCCAGACTGAAGCACAGAGAGANAAAAAAATGGAAAATACAGAAAAGAGCGTAAGAGACATATGGGACACGGTGAAAAGGTCTAACATACGTGTAATTGGAGTCCCAGAAGGAGAGGAGAGAGAGAATGGGGCAGAAGCAATATTTGAAGAGATAATGGCCGAGAATTTTCCAAAACTGATGAAAGACATCAANCCACAGATTCAAGAAGCTCTACGAACCCCAAGCAGGATAAATACAAAGAAAACCACACCTAGGCACATCATAGTAAAACTGCTGAAAACCAAAGACAAAGAGAAAATCTTAAAAGCAGCCAGAGAAAAAAGACACATTACCTTCAAAGGAGCAACAATAAGACTGACAGCTGACTTCTCAACAGAAACAATGGAAGCCAGAAGACAATGGAATGACATCTTTAAAGTGCTGAAAGAAAATAACTGCCAACCTAGAATTCTATACCCAGGAAAATATCCTTCAAAAATGAAGGCGAAATAAAGACATTTTCAGACAAACAAAAACTGAGAGAATTCGTCACCAGCAGACCTGCACTAAAAGAAATACTAAAGGAAGTTCTTCAGGCAGAAGGAAAATGATCCCAGATGGAAGCACGGAAATGCAGGAAGGAATGAAGAGCAACGGAAAGGGTAAATATGTGGGTAAATCTAAATGAATATTGACTGTATAAAACAATAATAATAATGTCTTGTGGGGTTTAAAATATATATAGAATTAAAATACATGACAACAATAGCACAAAAGGGAAGAGAGGGTAAATGGAGTTAAAGTGTTCTAAGGTCCTTGCATTGTCCGGGAAGTGGTAAAGTACTAATTTATATTAGACTTTAATAAGTCAAGGATGCATGTTGTAATCTCTAGGGTAACCACTAAAAGAATAGTAAAAGAATGTATAACTAACAAGCTAATAGAGAGGGAAAATGGAATAATAAAAATATTCGATTAATCCAAAAGAAGGCAAGAAAAAAGGAAAAAANAGAAAGAAAAACAGATGGGACAAATAGAAAACAAATAGTAAGATGGTAGATTTAAACCCAAATATATCAGTAATTACATTAAATGTAAATGGACTAAATACTCCAATTNAAAGANAAAGATTGTCAGACTGGATTTAAAAACAAAACCCAACTATATGCTGCTTACAAGAGACACACCTTAAATATAAGGAC
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| L1MEg_5end | FLI1 | 388 | 396 | + | 15.44 | ACCGGAAAT |
| L1MEg_5end | DOF4.2 | 1896 | 1906 | + | 15.42 | AAAAAGGAAAA |
| L1MEg_5end | CRF4 | 227 | 236 | - | 15.36 | CAGCCGCCGC |
| L1MEg_5end | ELK4 | 4 | 12 | + | 15.33 | CGCTTCCGG |
| L1MEg_5end | ERF105 | 227 | 236 | + | 15.30 | GCGGCGGCTG |
| L1MEg_5end | VIP1 | 1276 | 1285 | + | 15.30 | TGACAGCTGA |
| L1MEg_5end | ERF076 | 227 | 236 | - | 15.19 | CAGCCGCCGC |
| L1MEg_5end | Sox6 | 1313 | 1322 | - | 15.16 | CCATTGTCTT |
| L1MEg_5end | RAMOSA1 | 1031 | 1044 | + | 15.11 | AGAGGAGAGAGAGA |
| L1MEg_5end | ZIM3 | 1291 | 1301 | + | 15.08 | CAACAGAAACA |
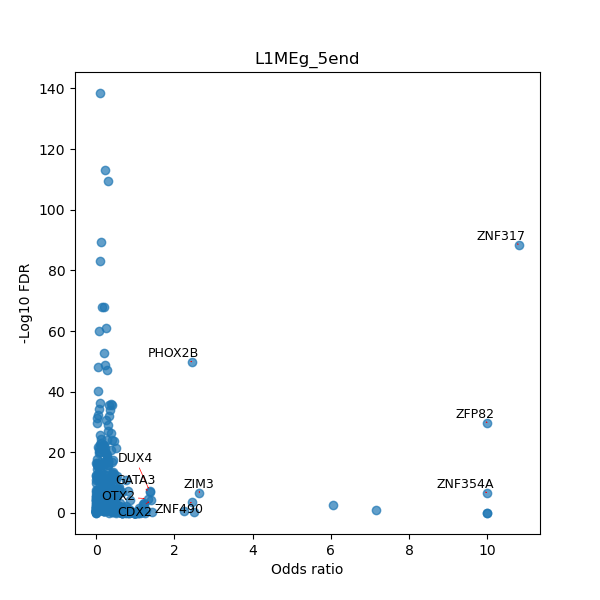
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.